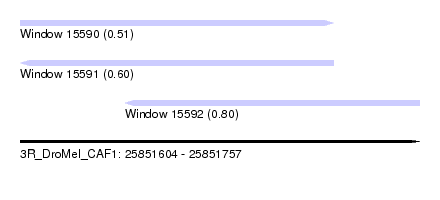
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 25,851,604 – 25,851,757 |
| Length | 153 |
| Max. P | 0.797144 |

| Location | 25,851,604 – 25,851,724 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -28.28 |
| Energy contribution | -29.88 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25851604 120 + 27905053 CGCGCCCAAGUGCAUCCUUCGGGUCCUUCGGUGUCCUUGUAAUGGGCGUUGCACCUGAAAAGUAGGCGCUUCCGCGCUGAACUUUUCCGCCUGUGCUGGCAACUUUUGCCGCGCACAAAG .(((((((..((((......(((..(....)..))).)))).))))))).((....(((((((.(((((....)))))..))))))).)).((((((((((.....))))).)))))... ( -47.50) >DroSec_CAF1 2827 120 + 1 CGCGCCCAAGUGCAUACUUCGGGUCCUUCGGUGUCCUUGCAAUGGGCGUUGCACCUGAAAAGUGGGCGCUUCCGCGCUGAACUUUUCCGCCUAUGCUGGCAACUUUUGCCGCGCACAAAG .(((((((..((((......(((..(....)..))).)))).)))))))(((....(((((((.(((((....)))))..))))))).......((.((((.....)))))))))..... ( -46.50) >DroSim_CAF1 2920 111 + 1 CGCGCCCAAGUGCAU---------CCUUCGGUGUCCUUGCAAUGGGCGUUGCACCUGAAAAGUGGGCGCUUCCGCGCUGAACUUUUCCGCCUAUGCUGGCAACUUUUGCCGCGCACAAAG .((((..(((..(((---------(....))))..)))(((.((((((........(((((((.(((((....)))))..)))))))))))))))).((((.....))))))))...... ( -43.90) >DroEre_CAF1 3413 111 + 1 CGCGCCCAAGUGCAU---------CCUUCGGUGUCCUUGCACUGGGCGUUGCACCUGAAAAGUGGGCGCUUCCGCGCUGAACUUUUCCGGCUUUGCUGGCAACUUUUGCCGCGUACAAAG .(((((((.(((((.---------((....).)....))))))))))))(((.((.(((((((.(((((....)))))..))))))).))....((.((((.....)))))))))..... ( -47.40) >DroAna_CAF1 1940 93 + 1 --------AGAGCCU---------CCUCCGAUGGCAUUGCAGUGGGCUUUGCACCUGAAAAGUGAGUGUUUCCGUGCUGAACUUU---------GCCGGCAACUUUU-CCGUAGACAAAG --------((((((.---------.((.((((...)))).))..))))))....(((....(.(((.(((.(((.((........---------))))).)))..))-)).)))...... ( -21.30) >consensus CGCGCCCAAGUGCAU_________CCUUCGGUGUCCUUGCAAUGGGCGUUGCACCUGAAAAGUGGGCGCUUCCGCGCUGAACUUUUCCGCCUAUGCUGGCAACUUUUGCCGCGCACAAAG .((((......((((..............(((((...(((.....)))..))))).(((((((.(((((....)))))..))))))).....)))).((((.....))))))))...... (-28.28 = -29.88 + 1.60)

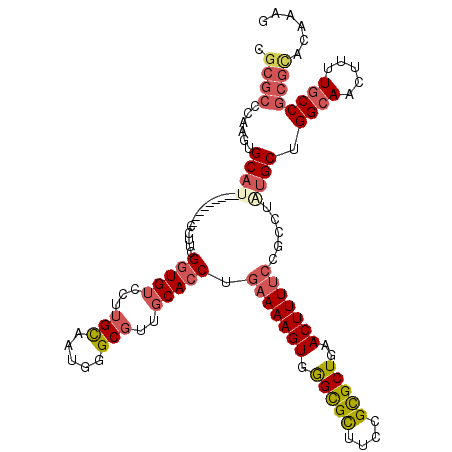
| Location | 25,851,604 – 25,851,724 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -26.98 |
| Energy contribution | -29.90 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 25851604 120 - 27905053 CUUUGUGCGCGGCAAAAGUUGCCAGCACAGGCGGAAAAGUUCAGCGCGGAAGCGCCUACUUUUCAGGUGCAACGCCCAUUACAAGGACACCGAAGGACCCGAAGGAUGCACUUGGGCGCG ..((((((..((((.....)))).))))))((((((((((...((((....))))..)))))))(((((((..(.((.......)).).((...(....)...)).)))))))...))). ( -47.70) >DroSec_CAF1 2827 120 - 1 CUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUUGCAAGGACACCGAAGGACCCGAAGUAUGCACUUGGGCGCG .....(((((((((.....)))).((....)).(((((((...((((....))))..)))))))..))))).((((((.((((...((..((.......))..)).))))..)))))).. ( -46.40) >DroSim_CAF1 2920 111 - 1 CUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUUGCAAGGACACCGAAGG---------AUGCACUUGGGCGCG .....(((((((((.....)))).((....)).(((((((...((((....))))..)))))))..))))).((((((.((((.(....)(....)---------.))))..)))))).. ( -44.00) >DroEre_CAF1 3413 111 - 1 CUUUGUACGCGGCAAAAGUUGCCAGCAAAGCCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAGUGCAAGGACACCGAAGG---------AUGCACUUGGGCGCG ..((((((((((((.....)))).)).......(((((((...((((....))))..)))))))..))))))(((((((((((.(....)(....)---------.))))).)))))).. ( -47.60) >DroAna_CAF1 1940 93 - 1 CUUUGUCUACGG-AAAAGUUGCCGGC---------AAAGUUCAGCACGGAAACACUCACUUUUCAGGUGCAAAGCCCACUGCAAUGCCAUCGGAGG---------AGGCUCU-------- ((((((...(((-........)))))---------))))...(((..(....).....((((((.((((((..((.....))..)).)))).))))---------)))))..-------- ( -22.70) >consensus CUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUUGCAAGGACACCGAAGG_________AUGCACUUGGGCGCG ..((((((((((((.....)))).)).......(((((((...((((....))))..)))))))..))))))((((((..(((.......................)))...)))))).. (-26.98 = -29.90 + 2.92)

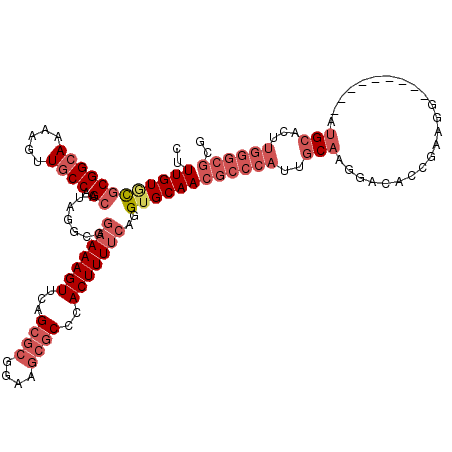
| Location | 25,851,644 – 25,851,757 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -30.04 |
| Energy contribution | -32.04 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.797144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
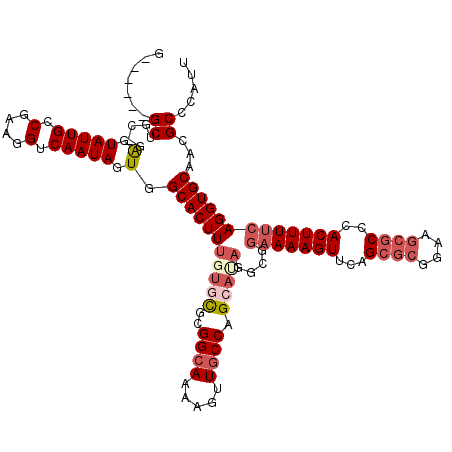
>3R_DroMel_CAF1 25851644 113 - 27905053 G------GCUGC-GAGUAUUGCCGAAGGUCAAUAGUGGCACUUUGUGCGCGGCAAAAGUUGCCAGCACAGGCGGAAAAGUUCAGCGCGGAAGCGCCUACUUUUCAGGUGCAACGCCCAUU (------(((((-(.....((((((((((((....)))).))))((((..((((.....)))).)))).))))(((((((...((((....))))..)))))))...))))..))).... ( -47.90) >DroSec_CAF1 2867 113 - 1 G------GCUGC-GAGUAUUGCCGAAGGUCAAUAGUGGCACUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUU (------((...-...(((((.(....).)))))((.(((((((((((..((((.....)))).)))))....(((((((...((((....))))..))))))))))))).))))).... ( -45.30) >DroSim_CAF1 2951 113 - 1 G------GCUGG-GAGUAUUGCCGAAGGUCAAUAGUGGCACUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUU .------..(((-(..(((((.(....).)))))((.(((((((((((..((((.....)))).)))))....(((((((...((((....))))..))))))))))))).)).)))).. ( -46.50) >DroEre_CAF1 3444 113 - 1 G------GCUGC-GAGUAUUGCCGAAGGUCAAUAGUGGCACUUUGUACGCGGCAAAAGUUGCCAGCAAAGCCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAGU (------(((((-.(.(((((.(....).))))).).)))..((((((((((((.....)))).)).......(((((((...((((....))))..)))))))..)))))).))).... ( -45.30) >DroAna_CAF1 1963 110 - 1 CGCCAGAGCUGCUGGUUAUUGCCGAAGGUCAAUAGUGGCACUUUGUCUACGG-AAAAGUUGCCGGC---------AAAGUUCAGCACGGAAACACUCACUUUUCAGGUGCAAAGCCCACU .(((.((.((..((((....)))).)).))......((((((((.((....)-))))).)))))))---------...(((..((((.((((........))))..))))..)))..... ( -33.00) >consensus G______GCUGC_GAGUAUUGCCGAAGGUCAAUAGUGGCACUUUGUGCGCGGCAAAAGUUGCCAGCAUAGGCGGAAAAGUUCAGCGCGGAAGCGCCCACUUUUCAGGUGCAACGCCCAUU .......((.....(.(((((.(....).))))).).(((((((((((..((((.....)))).)))))....(((((((...((((....))))..)))))))))))))...))..... (-30.04 = -32.04 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:41:35 2006