
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,118,761 – 3,118,918 |
| Length | 157 |
| Max. P | 0.999876 |

| Location | 3,118,761 – 3,118,863 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -37.46 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.98 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3118761 102 + 27905053 GCAGAUAAUUGCUGGGGACCGUCCUCCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUG ((((....)))).((((......))))(((((((((((((.....))))..(.(((((((....).))))))))))).(((((.....))))).)))))... ( -36.20) >DroSec_CAF1 6410 102 + 1 GCAGAUAAUUGCUGGGGCCGGUCCUCCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUG ((((....))))..((((((....(((((((((..(((((.....)))))..)).))))....(((....))))))..(((((.....)))))..)))))). ( -38.70) >DroSim_CAF1 11718 102 + 1 GCAGAUAAUUGCUGGGGCCGGUCCUCCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUG ((((....))))..((((((....(((((((((..(((((.....)))))..)).))))....(((....))))))..(((((.....)))))..)))))). ( -38.70) >DroEre_CAF1 6178 100 + 1 GCAGAUAAUUGCUGGC--CCGUCCUCCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACGGUGGCCUG ((((....)))).(((--(...(((((((((((..(((((.....)))))..)).))))....(((....))))))..(((((.....))))))).)))).. ( -38.10) >DroYak_CAF1 10092 102 + 1 GCAGAUAAUUGCUGCCCCCCGUCCACCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCACUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUG ((((.......)))).....(.((((.((((((..(((((.....)))))..)).))))..(((((....)))))...(((((.....))))).)))).).. ( -35.60) >consensus GCAGAUAAUUGCUGGGGCCCGUCCUCCGCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUG ((((....)))).((((......))))(((((((((((((.....))))..(.(((((((....).))))))))))..(((((.....)))))))))))... (-32.14 = -32.98 + 0.84)

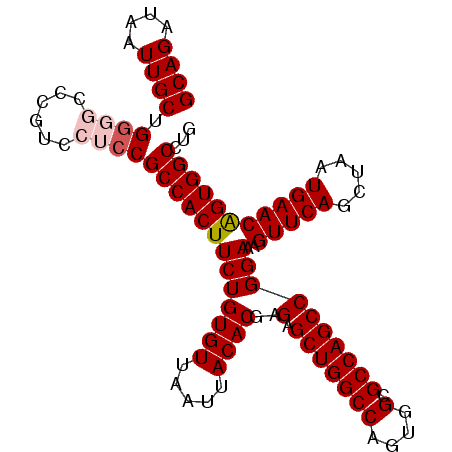
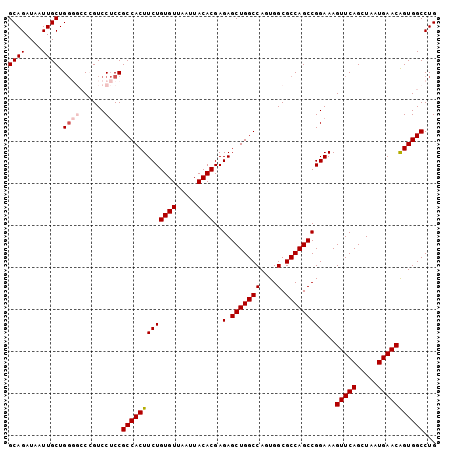
| Location | 3,118,788 – 3,118,900 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.68 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -37.54 |
| Energy contribution | -37.78 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3118788 112 + 27905053 GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGAAAGCUGAAGCUUUUCAGCGGCAUUUCAAAUGGACGA .(((.....((((.....))))((((((((((((.((((....))).....(((((.....)))))).))))).....((((((....))))))))).))))...))).... ( -38.40) >DroSec_CAF1 6437 112 + 1 GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGA .(((.....((((.....))))((((((((((((.((((....))).....(((((.....)))))).))))).....((((((....))))))))).))))...))).... ( -39.80) >DroSim_CAF1 11745 112 + 1 GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGA .(((.....((((.....))))((((((((((((.((((....))).....(((((.....)))))).))))).....((((((....))))))))).))))...))).... ( -39.80) >DroEre_CAF1 6203 112 + 1 GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACGGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGA .(((.....((((.....))))((((((((((((.((((....))).....(((((.....)))))).))))).....((((((....))))))))).))))...))).... ( -39.80) >DroYak_CAF1 10119 112 + 1 GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCACUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGAAAGCUAAAGCUCCUCAGCGGCAUUUCAAAUGGACGA (((...(((((((.....)))).)))(((((((((((((....))).....(((((.....)))))))))))).((.(((....)))..))))))))............... ( -40.10) >consensus GCCACUUCUGUGUUAAUUACACGAGAGCUGGCCAGUGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGA .(((.....((((.....))))((((((((((((.((((....))).....(((((.....)))))).))))).....((((((....))))))))).))))...))).... (-37.54 = -37.78 + 0.24)

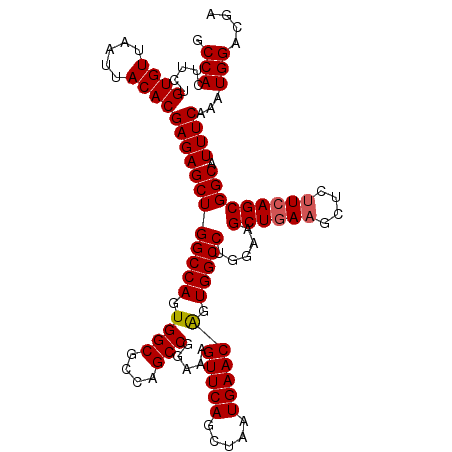
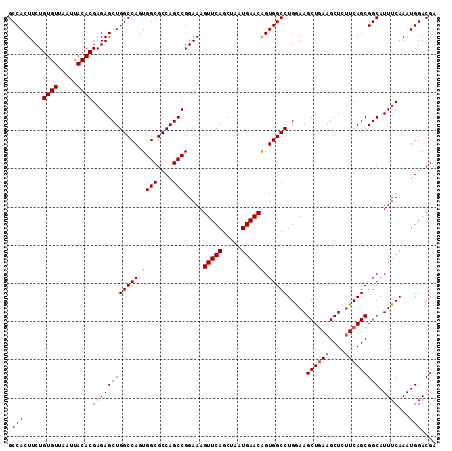
| Location | 3,118,788 – 3,118,900 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.68 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -35.94 |
| Energy contribution | -35.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3118788 112 - 27905053 UCGUCCAUUUGAAAUGCCGCUGAAAAGCUUCAGCUUUCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCACUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............(((((((.((((.(((((((..(((....)))......))))))))))).)))).)))((((((.......(((.((((.....))))))))))))) ( -37.51) >DroSec_CAF1 6437 112 - 1 UCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCACUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............((((((((.(((((((((((..(((....)))......)))))).......(((..(.....)..))))))))))((((.....))))...)))))) ( -37.40) >DroSim_CAF1 11745 112 - 1 UCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCACUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............((((((((.(((((((((((..(((....)))......)))))).......(((..(.....)..))))))))))((((.....))))...)))))) ( -37.40) >DroEre_CAF1 6203 112 - 1 UCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACCGUUCAUUAGCUGAACUUUCCGGCUGGCGCCACUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............((((((((.(((((((((((....(((....)))....)))))).......(((..(.....)..))))))))))((((.....))))...)))))) ( -36.00) >DroYak_CAF1 10119 112 - 1 UCGUCCAUUUGAAAUGCCGCUGAGGAGCUUUAGCUUUCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCAGUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............(((((((((((((....))))...((((((((..(.(((((((.......))))))).).))))))))....)))((((.....))))...)))))) ( -41.10) >consensus UCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCACUGGCCAGCUCUCGUGUAAUUAACACAGAAGUGGC ...............(((((((.((((.(((((((..(((....)))......))))))))))).).((((((.......))))))(((.((((.....))))))))))))) (-35.94 = -35.66 + -0.28)

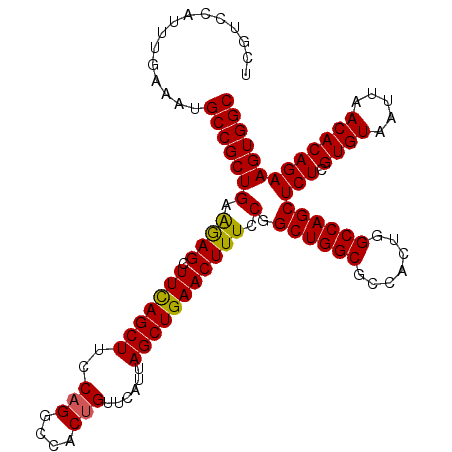
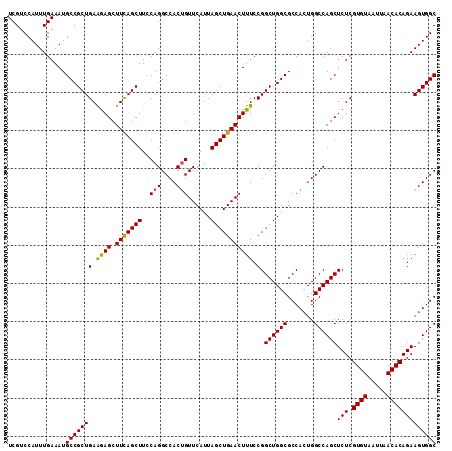
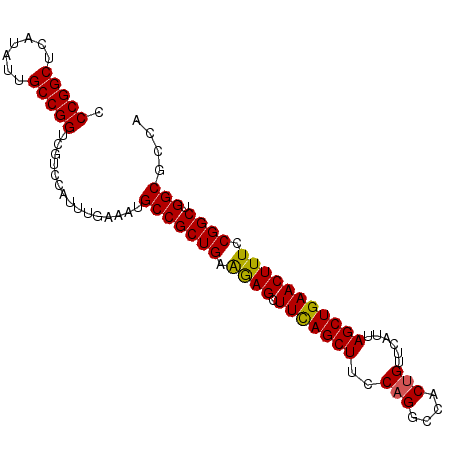
| Location | 3,118,823 – 3,118,918 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.68 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -33.04 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3118823 95 + 27905053 UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGAAAGCUGAAGCUUUUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ...(((((....(((((((((.....))))).((.((..((((((....))))))..)).)).))))...))).)).(((((.......))))). ( -34.40) >DroSec_CAF1 6472 95 + 1 UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ...(((((....(((((((((.....)))))....(((.....((((((....))))))))).))))...))).)).(((((.......))))). ( -33.40) >DroSim_CAF1 11780 95 + 1 UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ...(((((....(((((((((.....)))))....(((.....((((((....))))))))).))))...))).)).(((((.......))))). ( -33.40) >DroEre_CAF1 6238 95 + 1 UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACGGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ..((.(((((((....(((((.....)))))..))).))))..((((((....)))))).))...............(((((.......))))). ( -34.00) >DroYak_CAF1 10154 95 + 1 UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGAAAGCUAAAGCUCCUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ...(((((....(((((((((.....))))).((.((.(((.(((....)))..))))).)).))))...))).)).(((((.......))))). ( -32.50) >consensus UGGCGCCAGCCGGAAAGUUCAGCUAAUGAACAGUGGCCUGGAAGCUGAAGCUCUUCAGCGGCAUUUCAAAUGGACGACCGGCAAUAUGAGCCGGG ...(((((....(((((((((.....))))).((.((.(((((((....))).)))))).)).))))...))).)).(((((.......))))). (-33.04 = -33.00 + -0.04)

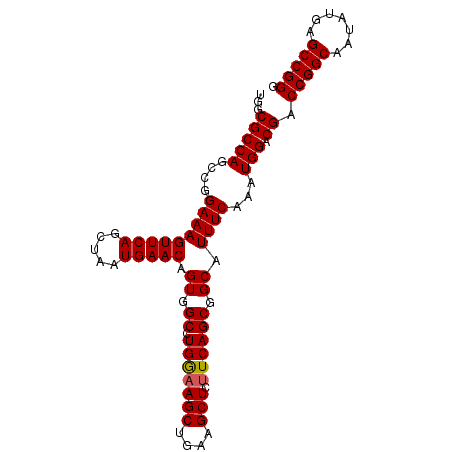
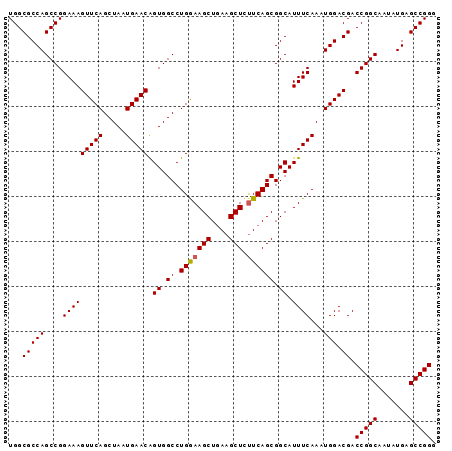
| Location | 3,118,823 – 3,118,918 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.68 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -33.54 |
| Energy contribution | -33.26 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3118823 95 - 27905053 CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAAAAGCUUCAGCUUUCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((.((((.(((((((..(((....)))......))))))))))).)))).))).... ( -34.90) >DroSec_CAF1 6472 95 - 1 CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((.((((.(((((((..(((....)))......))))))))))).)))).))).... ( -34.30) >DroSim_CAF1 11780 95 - 1 CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((.((((.(((((((..(((....)))......))))))))))).)))).))).... ( -34.30) >DroEre_CAF1 6238 95 - 1 CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACCGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((.((((.(((((((....(((....)))....))))))))))).)))).))).... ( -32.90) >DroYak_CAF1 10154 95 - 1 CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAGGAGCUUUAGCUUUCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((..(((.(((((((..(((....)))......))))))))))..)))).))).... ( -34.30) >consensus CCCGGCUCAUAUUGCCGGUCGUCCAUUUGAAAUGCCGCUGAAGAGCUUCAGCUUCCAGGCCACUGUUCAUUAGCUGAACUUUCCGGCUGGCGCCA .(((((.......)))))...............(((((((.((((.(((((((..(((....)))......))))))))))).)))).))).... (-33.54 = -33.26 + -0.28)

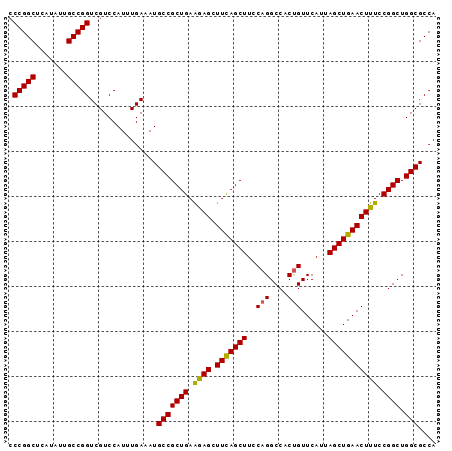
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:45 2006