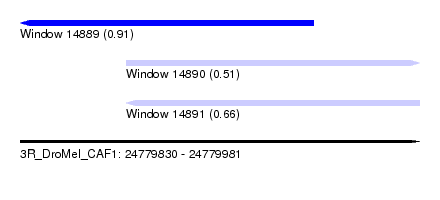
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 24,779,830 – 24,779,981 |
| Length | 151 |
| Max. P | 0.914559 |

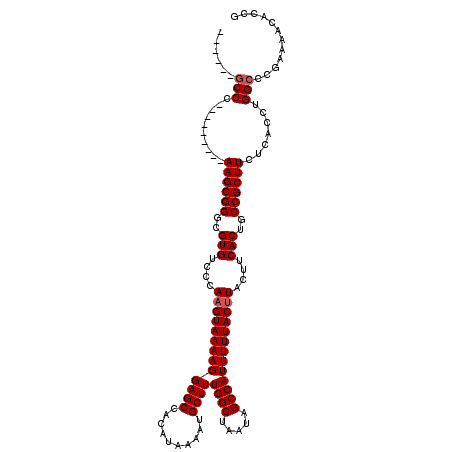
| Location | 24,779,830 – 24,779,941 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.87 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.79 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779830 111 - 27905053 CAAGCAGCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ...(((((((---------....))))(((....(((((((((((((..........))))((((.....)))))))))))))....)))))).(((........)))............ ( -27.50) >DroSec_CAF1 10921 105 - 1 ------GCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ------((((---------....))))((((...(((((((((((((..........))))((((.....)))))))))))))..(((...((((.........))))..))).)))).. ( -26.40) >DroSim_CAF1 6293 105 - 1 ------GCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ------((((---------....))))((((...(((((((((((((..........))))((((.....)))))))))))))..(((...((((.........))))..))).)))).. ( -26.40) >DroEre_CAF1 19907 114 - 1 ------GCCCAAACGGGCCAAGCGGGCGUGUCCCAUGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ------.......((((((((((((..(((......(((((((((((..........))))((((.....)))))))))))......)))..))))).......)))))))......... ( -35.41) >DroYak_CAF1 11516 105 - 1 ------ACCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ------....---------((((((..(((....(((((((((((((..........))))((((.....)))))))))))))....)))..))))))...................... ( -24.40) >consensus ______GCCC_________AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUACUUACUUCACUGCCGCUUCUCACCUGGCCCGAAAACACCG ......(((..........((((((..(((....(((((((((((((..........))))((((.....)))))))))))))....)))..)))))).......)))............ (-25.39 = -25.79 + 0.40)


| Location | 24,779,870 – 24,779,981 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779870 111 + 27905053 UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACUUGGGACACGCCCGCUU---------GGGCUGCUUGGGCUGCUUGGCCUGCAUGCUCUGCAGGAUCCCUCGGAUCC .......(((((.(((((.(((((......(((..(((........))).)))((((....---------))))))))).)))))..)))))((((.....))))((((((...)))))) ( -43.50) >DroSec_CAF1 10961 102 + 1 UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACUUGGGACACGCCCGCUU---------GGGC---------UGCUUGGCCUGCAUGCUCCGCAGGAUCCCUCGGAUCC .......(((((.(((((.((((.......((((......))))..(((.....)))))))---------.)))---------))..)))))(((.......)))((((((...)))))) ( -32.90) >DroSim_CAF1 6333 100 + 1 UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACUUGGGACACGCCCGCUU---------GGGC---------UGCUUGGCCUGCAUGCUCCGCAGUAUCCCUCCGCA-- ........((.....(((((.(((......(((..(((........))).)))((((....---------))))---------))))))))((((.......)))).........)).-- ( -29.00) >DroEre_CAF1 19947 111 + 1 UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACAUGGGACACGCCCGCUUGGCCCGUUUGGGC---------UGCUUGGCCUGCAUGCUCUGCAGGAUCCCUGGGAUCC ......((((.....))))..(((...(((((((((..........(((.....)))((..(((((....))))---------)))..)))).)))))...))).((((((...)))))) ( -38.00) >DroYak_CAF1 11556 102 + 1 UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACUUGGGACACGCCCGCUU---------GGGU---------UGUUUGGCCUGCAUGCCCCGUAGGAGUCCUCGGAUCC .......(((.....((((((.((......(((..(((........))).)))((((....---------))))---------))))))))......)))(((.(((...)))))).... ( -27.60) >consensus UAAAAAUGGCUAUUAGCCAAAGCAUUUUAUGUGGCUCCUUUUACUUGGGACACGCCCGCUU_________GGGC_________UGCUUGGCCUGCAUGCUCCGCAGGAUCCCUCGGAUCC ......((((.....))))(((((......(((..(((........))).)))((((.............)))).........)))))(((((((.......)))))...))........ (-26.80 = -26.52 + -0.28)

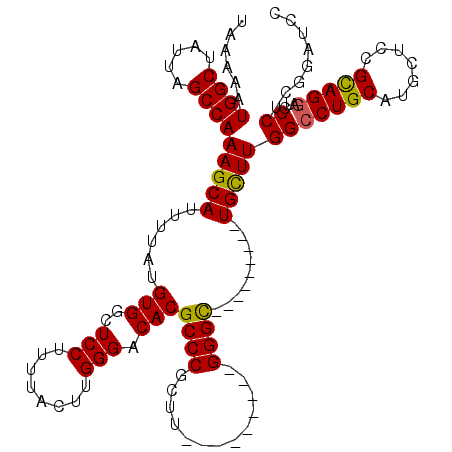
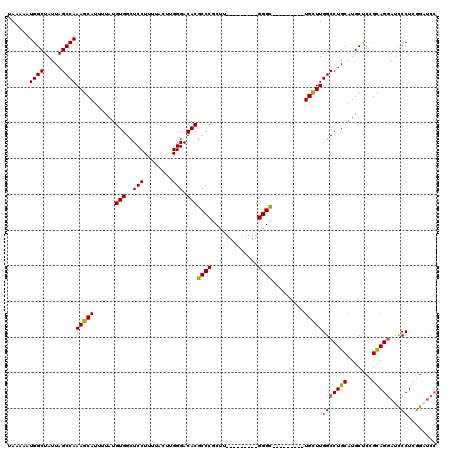
| Location | 24,779,870 – 24,779,981 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 24779870 111 - 27905053 GGAUCCGAGGGAUCCUGCAGAGCAUGCAGGCCAAGCAGCCCAAGCAGCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA ((((((...))))))((((.....))))(((.....((((.(((((((((---------....))))(((.(((........)).).)))......))))).))))....)))....... ( -41.10) >DroSec_CAF1 10961 102 - 1 GGAUCCGAGGGAUCCUGCGGAGCAUGCAGGCCAAGCA---------GCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA ((((((...))))))((((.....))))(((((((((---------((((---------....))))(((.(((........)).).)))......))).)))))).............. ( -35.10) >DroSim_CAF1 6333 100 - 1 --UGCGGAGGGAUACUGCGGAGCAUGCAGGCCAAGCA---------GCCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA --(((...((((((((((...((......))...)))---------((((---------....)))))))))))..)))...(((((..........)))))(((.....)))....... ( -31.90) >DroEre_CAF1 19947 111 - 1 GGAUCCCAGGGAUCCUGCAGAGCAUGCAGGCCAAGCA---------GCCCAAACGGGCCAAGCGGGCGUGUCCCAUGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA ((((((...))))))...(((((((....(((..((.---------((((....))))...)).)))(((.(((........)).).))).....)))))))(((.....)))....... ( -37.40) >DroYak_CAF1 11556 102 - 1 GGAUCCGAGGACUCCUACGGGGCAUGCAGGCCAAACA---------ACCC---------AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA ........((.((((((((((((((((..((......---------....---------..))..)))))))))..))...))))))).............((((.....))))...... ( -31.70) >consensus GGAUCCGAGGGAUCCUGCGGAGCAUGCAGGCCAAGCA_________GCCC_________AAGCGGGCGUGUCCCAAGUAAAAGGAGCCACAUAAAAUGCUUUGGCUAAUAGCCAUUUUUA ........((....(((((.....)))))((((((((.........((((.............))))(((.(((........)).).)))......))).)))))......))....... (-24.60 = -24.96 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:31:22 2006