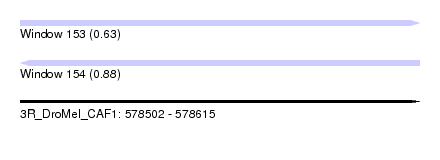
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 578,502 – 578,615 |
| Length | 113 |
| Max. P | 0.877793 |

| Location | 578,502 – 578,615 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -17.42 |
| Energy contribution | -17.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
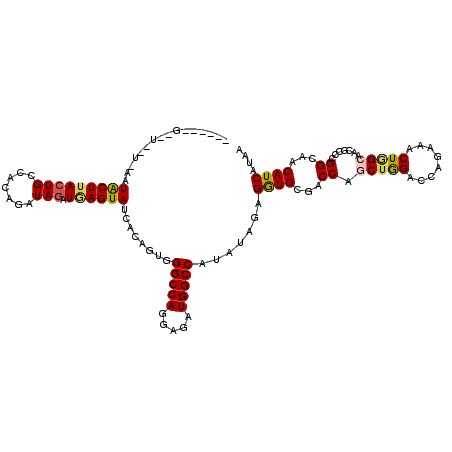
>3R_DroMel_CAF1 578502 113 + 27905053 ------G-UUUUUGAUUACUCACUGCCACAGAUAGAUUAGUAUCACAGUUGGCCACGAGAUGGCCAUGUAGAGGUGGGACCAGCUGGACCAGAAGCUGGCAACUCCGGGCAACAUCAUUA ------.-....((((..((((((.(.(((((((......)))).....((((((.....))))))))).).)))))).((((((........))))))........(....)))))... ( -31.70) >DroVir_CAF1 1233 118 + 1 CAAUAAGACU--UUGAUACUUACUGCCACAGGUAGAUGAGUAUGACUGUGGGCCAGGAGAUGGCCAUAUAGAGGUGAGACCAGCUGGACCAAAAACUGGCAACGCCUGGCAACAUCAUAA ..........--...((((((((((((...))))).)))))))..((((((((((.....))))).))))).((((...((((.((..(((.....)))...)).))))...)))).... ( -36.00) >DroGri_CAF1 1126 113 + 1 CAGAAAG-------GUUACUUACUGCCACAAAUAGAUGAGUAUCACUGUGGGCCAGGAGAUGGCCAUAUAGAGGUGCGACCAACUGGACCAGAAACUAGCCACACCCGGCAACAUCAUAA (((.(((-------....))).))).........(((((((.((.((((((((((.....))))).))))).(((.((......)).))).)).))).(((......)))..)))).... ( -33.30) >DroWil_CAF1 434 113 + 1 ------G-AUUUUCCAUGCUUACUGCCAUAGAUAGAUAAGUAUUACAGUGGGCCAUGAAAUGGCCAUAUAAAGAUGUGACCAACUGGACCAGAAACUUGCAACGCCCGGCAACAUCAUGA ------.-......((((.....((((...((((......)))).....((((.......(((.(((((....))))).))).(((...)))...........))))))))....)))). ( -22.80) >DroMoj_CAF1 1195 108 + 1 ------------UUAAUACUUACUGCCAAAGGUAAAUGAGUAUUACUGUGGGCCAAGAAAUGGCCAUAUAAAGAUGCGACCAGCUAGACCAAAAGCUGGCCAAUCCUGGUAACAUCAUAA ------------.(((((((((.((((...))))..))))))))).(((((((((.....))))).))))..((((..(((((((((........)))))......))))..)))).... ( -31.30) >DroAna_CAF1 1042 108 + 1 ------A-UU-----CUACUUACUGCCACAGAUAGAUAAGUAUCACAGUUGGCCAGGAGAUGGCCAUGUAGAGGUGGGACCAGCUGGAAAAGAAGCUAGCCAGCCCAGGAAGCAUCAUAA ------.-((-----((((((.((((....((((......)))).....((((((.....)))))).))))))))))))((.(((((............)))))...))........... ( -30.70) >consensus ______G__U__U_AAUACUUACUGCCACAGAUAGAUGAGUAUCACAGUGGGCCAGGAGAUGGCCAUAUAGAGGUGCGACCAGCUGGACCAGAAACUGGCAACGCCCGGCAACAUCAUAA ................(((((((((.......))).))))))........(((((.....))))).......((((...((.(((((........))))).......))...)))).... (-17.42 = -17.42 + 0.00)


| Location | 578,502 – 578,615 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -20.57 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
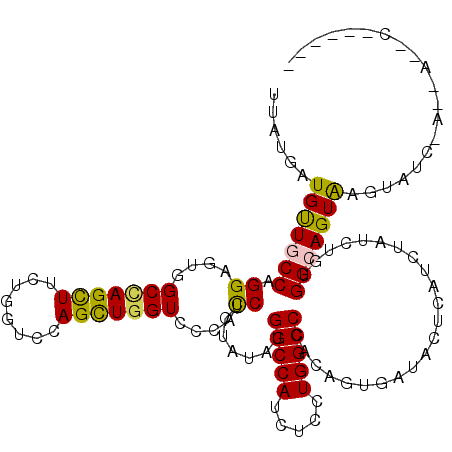
>3R_DroMel_CAF1 578502 113 - 27905053 UAAUGAUGUUGCCCGGAGUUGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCGUGGCCAACUGUGAUACUAAUCUAUCUGUGGCAGUGAGUAAUCAAAAA-C------ ........((((((((((..(((((((........)))))))..).....(((((((((((...)))))))..))))...........)))).)))))(((....)))....-.------ ( -35.70) >DroVir_CAF1 1233 118 - 1 UUAUGAUGUUGCCAGGCGUUGCCAGUUUUUGGUCCAGCUGGUCUCACCUCUAUAUGGCCAUCUCCUGGCCCACAGUCAUACUCAUCUACCUGUGGCAGUAAGUAUCAA--AGUCUUAUUG ..........((((((.(..((((......(((((....))....)))......))))...).))))))((((((..............))))))(((((((......--...))))))) ( -31.84) >DroGri_CAF1 1126 113 - 1 UUAUGAUGUUGCCGGGUGUGGCUAGUUUCUGGUCCAGUUGGUCGCACCUCUAUAUGGCCAUCUCCUGGCCCACAGUGAUACUCAUCUAUUUGUGGCAGUAAGUAAC-------CUUUCUG ..........((((((.(((((((......(((((....))....)))......)))))))..))))))((((((.(((....)))...))))))(((.(((....-------))).))) ( -33.70) >DroWil_CAF1 434 113 - 1 UCAUGAUGUUGCCGGGCGUUGCAAGUUUCUGGUCCAGUUGGUCACAUCUUUAUAUGGCCAUUUCAUGGCCCACUGUAAUACUUAUCUAUCUAUGGCAGUAAGCAUGGAAAAU-C------ (((((...((((((((..(((((.((....((.(((..((((((.((....)).)))))).....))))).)))))))..))..........))))))....))))).....-.------ ( -25.60) >DroMoj_CAF1 1195 108 - 1 UUAUGAUGUUACCAGGAUUGGCCAGCUUUUGGUCUAGCUGGUCGCAUCUUUAUAUGGCCAUUUCUUGGCCCACAGUAAUACUCAUUUACCUUUGGCAGUAAGUAUUAA------------ ....((((((((.((((((((((((((........))))))))).))))).....(((((.....)))))....)))))).))((((((........)))))).....------------ ( -32.50) >DroAna_CAF1 1042 108 - 1 UUAUGAUGCUUCCUGGGCUGGCUAGCUUCUUUUCCAGCUGGUCCCACCUCUACAUGGCCAUCUCCUGGCCAACUGUGAUACUUAUCUAUCUGUGGCAGUAAGUAG-----AA-U------ .............((((...(((((((........)))))))))))..(((((.((((((.....))))))(((((.(((..........))).)))))..))))-----).-.------ ( -33.90) >consensus UUAUGAUGUUGCCAGGAGUGGCCAGCUUCUGGUCCAGCUGGUCCCACCUCUAUAUGGCCAUCUCCUGGCCCACAGUGAUACUCAUCUAUCUGUGGCAGUAAGUAUC_A__A__C______ ......(((((((.((....(((((((........)))))))....)).......(((((.....))))).......................))))))).................... (-20.57 = -19.93 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:50 2006