
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 526,275 – 526,399 |
| Length | 124 |
| Max. P | 0.990618 |

| Location | 526,275 – 526,372 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -13.38 |
| Consensus MFE | -12.41 |
| Energy contribution | -11.10 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
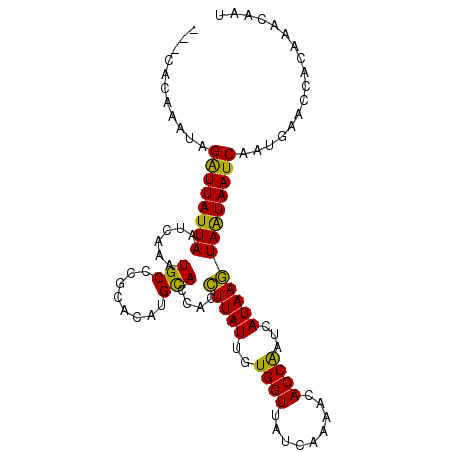
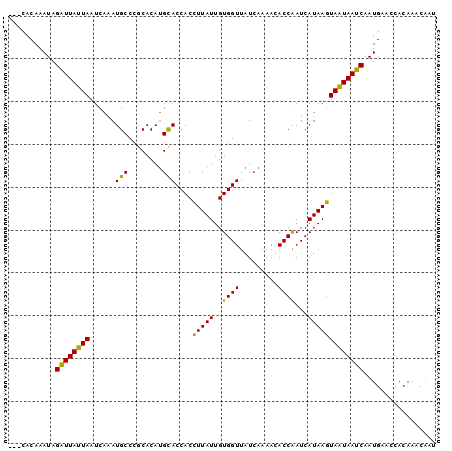
>3R_DroMel_CAF1 526275 97 + 27905053 ---CACAAAUAGAUUAUUAAUCAAAUGCCCGCACAUGCACCACCUUAUUGUGGUUAUCAAAAUACCAAUCAUAAGUAAUAAUCAAUGCACCACAAACAAU ---........((((((((...........((....)).....(((((..((((.........))))...)))))))))))))................. ( -14.10) >DroSec_CAF1 3151 97 + 1 ---CACAAAUAGGUUAUUAAUCAAAUGCCCGCACAUGUACCACCUUAUUGUGGUUAUCAAAACACCGAUCAUAAGUAAUAAUCAAUGAACCACAAACAAU ---........((((((((.....(((..((....((.(((((......)))))...))......))..)))...))))))))................. ( -12.40) >DroSim_CAF1 2513 96 + 1 ---CACAAAUAGGUUAUUAAUCAAAUGCCCGCACAUGUACCACCUUAUUGUGGUUAUCAAAACACCGAUCAUAAGUAAUAAUCAAUGA-CCACAAACAAU ---........(((((((.................((.(((((......)))))...)).......(((.((.....)).))))))))-))......... ( -13.30) >DroEre_CAF1 3454 100 + 1 AAACACAAAUAGAUUAUUAGUCAAAUGCCCACACAAGCACCACUUUAUUGUGGUUAUCAAAAUACCAAUCAUAAAUAGUAAUCAAUGAAUCACAAACUAU ...........((((((((.......((........))(((((......))))).....................))))))))................. ( -13.70) >consensus ___CACAAAUAGAUUAUUAAUCAAAUGCCCGCACAUGCACCACCUUAUUGUGGUUAUCAAAACACCAAUCAUAAGUAAUAAUCAAUGAACCACAAACAAU ...........((((((((......(((........)))....(((((..((((.........))))...)))))))))))))................. (-12.41 = -11.10 + -1.31)

| Location | 526,307 – 526,399 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -13.73 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
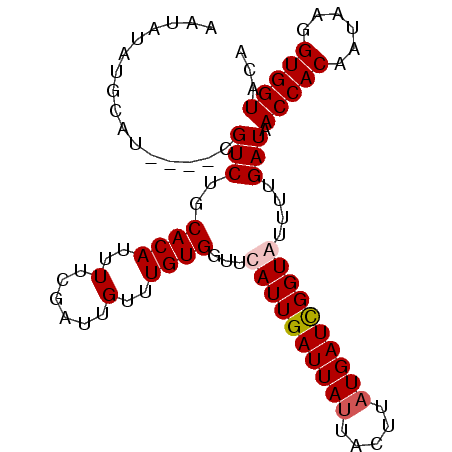
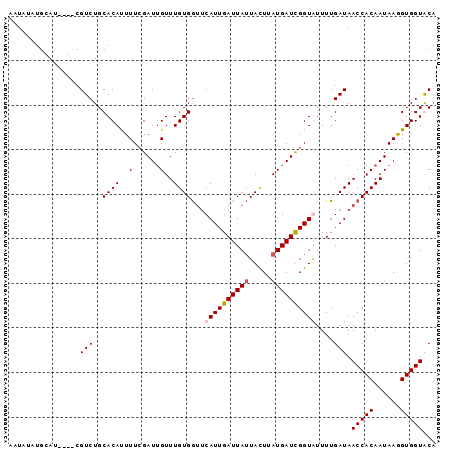
>3R_DroMel_CAF1 526307 92 - 27905053 AAUAUUUGUAU----CGUCUGCACAUUUUCGAUUGUUUGUGGUGCAUUGAUUAUUACUUAUGAUUGGUAUUUUGAUAACCACAAUAAGGUGGUGCA ......(((((----(..((..(((........)))(((((((..((..(....((((.......))))..)..)).)))))))..))..)))))) ( -20.80) >DroSec_CAF1 3183 96 - 1 AAUAUAUGCAUCUGCCGUCUGCACAUUUUCGAUUGUUUGUGGUUCAUUGAUUAUUACUUAUGAUCGGUGUUUUGAUAACCACAAUAAGGUGGUACA ............((((..((..(((........)))((((((((((((((((((.....)))))))))).......))))))))..))..)))).. ( -23.41) >DroSim_CAF1 2545 95 - 1 AAUAUAUGCAUCUGCCGUCUGCACAUUUUCGAUUGUUUGUGG-UCAUUGAUUAUUACUUAUGAUCGGUGUUUUGAUAACCACAAUAAGGUGGUACA ............((((..((..(((........)))((((((-(((((((((((.....))))))))))........)))))))..))..)))).. ( -22.40) >DroEre_CAF1 3489 80 - 1 ----------------GUCUGCACACUUUAGAUAGUUUGUGAUUCAUUGAUUACUAUUUAUGAUUGGUAUUUUGAUAACCACAAUAAAGUGGUGCU ----------------....((((((((((......(((((.((.((..(.(((((........)))))..)..)))).))))))))))).)))). ( -17.20) >consensus AAUAUAUGCAU____CGUCUGCACAUUUUCGAUUGUUUGUGGUUCAUUGAUUAUUACUUAUGAUCGGUAUUUUGAUAACCACAAUAAGGUGGUACA ................(((..((((..(......)..))))...((((((((((.....))))))))))....))).(((((......)))))... (-13.73 = -14.23 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:42 2006