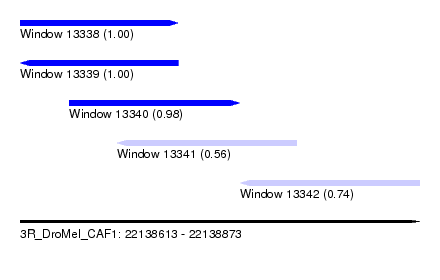
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,138,613 – 22,138,873 |
| Length | 260 |
| Max. P | 0.999987 |

| Location | 22,138,613 – 22,138,716 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22138613 103 + 27905053 UAUAUAUCUGUAUAUAUGU--------AGAAAAAAGUAAAAG-AGA--------AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG (((((((......))))))--------)((((((.(((((((-..(--------((.((((((((..........((((...))))..)))))))).))).....))))))).)))))). ( -28.50) >DroSec_CAF1 68942 103 + 1 UAUAUAUCUAUAUAUAUGU--------AGAAAAAAGUAAAAG-AGA--------AGGGGCGUUGCAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG (((((((......))))))--------)((((((.(((((((-...--------....(((((((............)))))))(((((....))))).......))))))).)))))). ( -30.40) >DroSim_CAF1 61926 103 + 1 UAUAUAUCUAUAUAUAUGU--------AGAAAAAAGUAAAAG-AGA--------AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG (((((((......))))))--------)((((((.(((((((-..(--------((.((((((((..........((((...))))..)))))))).))).....))))))).)))))). ( -28.50) >DroEre_CAF1 64110 102 + 1 UAUAUAUUUAUAUAUAUGU---------GGAAAAAGUGAAAG-AGA--------AGGGGCGUUGGGAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG (((((((......))))))---------)(((((((((((((-...--------....((((((.(..........).))))))(((((....))))).......))))))).)))))). ( -28.80) >DroYak_CAF1 61498 120 + 1 UAUAUAUUUAUAUAUACGAGUAUAUCUAGAAAAAAGUAAAAGGAGAAGCGGCGAUGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG .((((((((........))))))))...((((((.(((((((...((((.(((.((..(((((...........)))))..)))))(((....))))))).....))))))).)))))). ( -31.90) >consensus UAUAUAUCUAUAUAUAUGU________AGAAAAAAGUAAAAG_AGA________AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUG (((((((....)))))))..........((((((.((((((.................((((((..............))))))(((((....)))))........)))))).)))))). (-25.58 = -25.46 + -0.12)


| Location | 22,138,613 – 22,138,716 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.46 |
| SVM RNA-class probability | 0.999987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22138613 103 - 27905053 CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCCU--------UCU-CUUUUACUUUUUUCU--------ACAUAUAUACAGAUAUAUA .((((((.((((((........(((((....)))))((((((..............))))))....--------...-.))))))))))))..--------..((((((....)))))). ( -22.34) >DroSec_CAF1 68942 103 - 1 CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUGCAACGCCCCU--------UCU-CUUUUACUUUUUUCU--------ACAUAUAUAUAGAUAUAUA .((((((.((((((........(((((....)))))((((((((..........))))))))....--------...-.))))))))))))..--------..((((((....)))))). ( -27.00) >DroSim_CAF1 61926 103 - 1 CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCCU--------UCU-CUUUUACUUUUUUCU--------ACAUAUAUAUAGAUAUAUA .((((((.((((((........(((((....)))))((((((..............))))))....--------...-.))))))))))))..--------..((((((....)))))). ( -22.34) >DroEre_CAF1 64110 102 - 1 CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUCCCAACGCCCCU--------UCU-CUUUCACUUUUUCC---------ACAUAUAUAUAAAUAUAUA ......((((............(((((....)))))((((((..............))))))....--------...-..............---------))))............... ( -18.04) >DroYak_CAF1 61498 120 - 1 CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCCAUCGCCGCUUCUCCUUUUACUUUUUUCUAGAUAUACUCGUAUAUAUAAAUAUAUA .((((((.((((((........(((((....)))))((((((..............)))))).................))))))))))))............((((((....)))))). ( -22.54) >consensus CAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCCU________UCU_CUUUUACUUUUUUCU________ACAUAUAUAUAGAUAUAUA .((((((.((((((........(((((....)))))((((((..............)))))).................))))))))))))............((((((....)))))). (-21.76 = -22.00 + 0.24)


| Location | 22,138,645 – 22,138,756 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -35.60 |
| Energy contribution | -35.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22138645 111 + 27905053 AG-AGA--------AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA ..-...--------((((((((((..............))))))(((((....)))))......................))))((((((((..((......))..))))))))...... ( -36.24) >DroSec_CAF1 68974 111 + 1 AG-AGA--------AGGGGCGUUGCAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA ..-...--------(((((((((((............)))))))(((((....)))))......................))))((((((((..((......))..))))))))...... ( -39.90) >DroSim_CAF1 61958 111 + 1 AG-AGA--------AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA ..-...--------((((((((((..............))))))(((((....)))))......................))))((((((((..((......))..))))))))...... ( -36.24) >DroEre_CAF1 64141 111 + 1 AG-AGA--------AGGGGCGUUGGGAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA ..-...--------((((((((((.(..........).))))))(((((....)))))......................))))((((((((..((......))..))))))))...... ( -38.20) >DroYak_CAF1 61538 120 + 1 AGGAGAAGCGGCGAUGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA .((..((((((((((((((((.((((((((.....(((...)))(((((....)))))......)))))).)).....)))))))........)))).)))))..))............. ( -38.60) >consensus AG_AGA________AGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACAUUUUUUGCCCUGGUCUUUGUUCGCUCGCUUGGCCAGAGACCCAGAGA ...............((((((...((((((..(((((((...))(((((....)))))......)))))...))))))))))))((((((((..((......))..))))))))...... (-35.60 = -35.64 + 0.04)

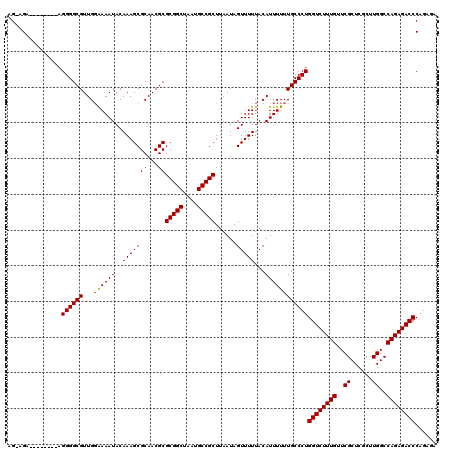
| Location | 22,138,676 – 22,138,793 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.98 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22138676 117 - 27905053 UGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU ..((---(..((((((.......))))))..))).((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. ( -36.60) >DroSec_CAF1 69005 117 - 1 UGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU ..((---(..((((((.......))))))..))).((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. ( -36.60) >DroSim_CAF1 61989 117 - 1 UGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU ..((---(..((((((.......))))))..))).((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. ( -36.60) >DroEre_CAF1 64172 117 - 1 UGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU ..((---(..((((((.......))))))..))).((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. ( -36.60) >DroYak_CAF1 61578 120 - 1 UGGGCGGCAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU .....(((..((((((.......))))))..))).((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. ( -36.90) >consensus UGGG___CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUGUCUCUGGGUCUCUGGCCAAGCGAGCGAACAAAGACCAGGGCAAAAAAUGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGU ..........((((((.......))))))......((.((((.((((.(((.((..(........)..)).)))))))))))....................(((((....))))))).. (-32.50 = -32.50 + 0.00)

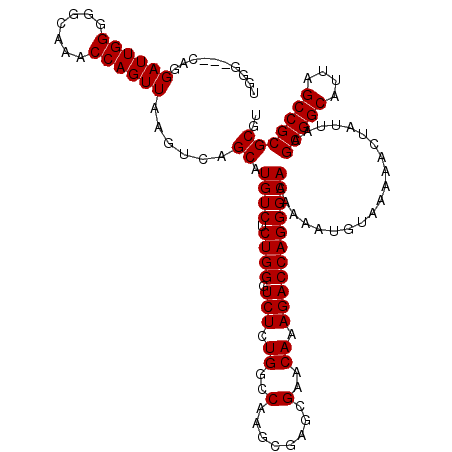
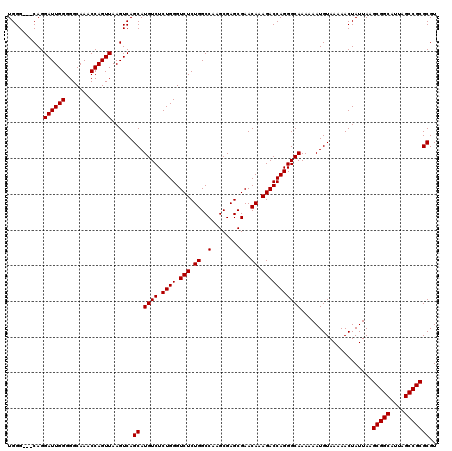
| Location | 22,138,756 – 22,138,873 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.45 |
| Mean single sequence MFE | -37.06 |
| Consensus MFE | -36.14 |
| Energy contribution | -36.62 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22138756 117 - 27905053 UCAAUGGGAUUUACAUUUUCCAGCUAUUGAACACGGUCGCUGGGUCUUAAAGCAUCGGACGGUAGUCGGAGUUUCAAUCCUGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG ...(((.(((((((.....(((((.((((....)))).)))))(((((.((((.((.(((....))).))))))(((((((...---.))))))))))))......).))))))..))). ( -35.60) >DroSec_CAF1 69085 117 - 1 UCAAUGGGAUUUACAUUUUCCAGCUAUUGAACACGGUGGCUGGGUCUUAAAGCAUCGGACGGUAGUCGGAGUUUCAAUGCUGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG ....((..((........(((((((((((....)))))))))))..........((.(((....))).))))..))(((((((.---(..((((((.......))))))..)))))))). ( -38.80) >DroSim_CAF1 62069 117 - 1 UCAAUGGGAUUUACAUUUUCCAGCUAUUGAACACGGUGGCUGGGUCUUAAAGCAUCGGACGGUAGUCGGAGUUUCAAUCCUGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG ...(((.(((((((.....((((((((((....))))))))))(((((.((((.((.(((....))).))))))(((((((...---.))))))))))))......).))))))..))). ( -39.80) >DroEre_CAF1 64252 117 - 1 UCAAUGGGAUUUACAUUUUCCAGCUAUUGAACACGGUCGCUGGGUCUUAAAGCAUCGGACGGUAGUCGGAGUUUCAAUCCUGGG---CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG ...(((.(((((((.....(((((.((((....)))).)))))(((((.((((.((.(((....))).))))))(((((((...---.))))))))))))......).))))))..))). ( -35.60) >DroYak_CAF1 61658 120 - 1 UCAAUGAGAUUUACAUUUUCCAGCUAUUGAACACGGUGGCUGGCUCUUAAAGCAUCGGAUGGUAGACGGAGUUUCAGUCCUGGGCGGCAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG .......(((((((......(((((((((....)))))))))(((((..((((..((.........))..))))((((((((.....)))))))))))))......).))))))...... ( -35.50) >consensus UCAAUGGGAUUUACAUUUUCCAGCUAUUGAACACGGUGGCUGGGUCUUAAAGCAUCGGACGGUAGUCGGAGUUUCAAUCCUGGG___CAGGAUUGGGGGCAAACCAGUUAAGUCAGCAUG ...(((.(((((((.....((((((((((....))))))))))........((.((.(((....))).)).(..((((((((.....))))))))..)))......).))))))..))). (-36.14 = -36.62 + 0.48)


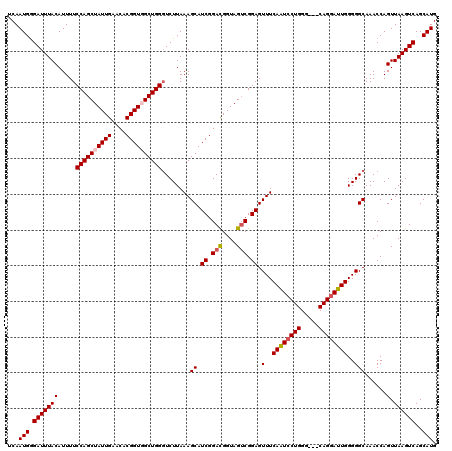
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:29 2006