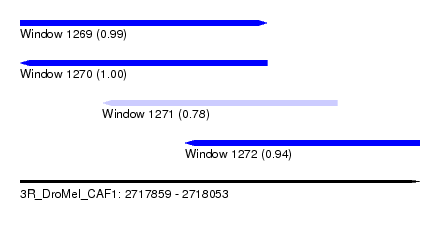
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,717,859 – 2,718,053 |
| Length | 194 |
| Max. P | 0.999967 |

| Location | 2,717,859 – 2,717,979 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2717859 120 + 27905053 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUCGGCUUCUGGAGAGUCGAGUAUCUCGUUCUCCCGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...((((.((((.((.....)).))))........((((((.(((.....))).))))...((((((.((((...)))))))))).))....))))(.(((((........))))).).. ( -31.30) >DroSec_CAF1 28133 120 + 1 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGGCUUCUGGAGAGUCGAGUAUCUCGUUCUCCCGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...((((.((((.((.....)).))))........(((((.(((........))))))...((((((.((((...)))))))))).))....))))(.(((((........))))).).. ( -31.60) >DroSim_CAF1 29321 120 + 1 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGGCUUCUGGAGAGUCGAGUAUCUCGUUCUCCCGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...((((.((((.((.....)).))))........(((((.(((........))))))...((((((.((((...)))))))))).))....))))(.(((((........))))).).. ( -31.60) >DroEre_CAF1 29211 120 + 1 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGGCUUCUGGAGAGUCGAGUAUCUCGUUCUACCGCUUUUCGUCGCUGGCCUUUCUUAUGGCCAACAG ...(((..((((.((.....)).))))........(((((.(((........))))))...((((((.((((...)))))))).)))).....)))(.(((((........))))).).. ( -28.40) >DroYak_CAF1 28930 120 + 1 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGGCUUCUGGAGAGUCGAGUAUCUCGUUCUCCCGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...((((.((((.((.....)).))))........(((((.(((........))))))...((((((.((((...)))))))))).))....))))(.(((((........))))).).. ( -31.60) >DroAna_CAF1 28034 120 + 1 AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGACUUCUGGUGAGUCGAGUAUCUCGUUUUCCAGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...((((...((...(((((((((((((....((...((((........)))).)).....)))))))))))))..))((......))....))))(.(((((........))))).).. ( -26.50) >consensus AAUGACACGUGAUUUUAUUUGAUUUACUUACAUCUGCAGCUCAAUUUUCAGUUUGGCUUCUGGAGAGUCGAGUAUCUCGUUCUCCCGCUUUUUGUCGCUGGCCUUUCUUAUGGCCAACAG ...(((..((((.((.....)).))))........((((((.(((.....))).))))...((((((.((((...)))))))))).)).....)))(.(((((........))))).).. (-27.72 = -28.08 + 0.36)


| Location | 2,717,859 – 2,717,979 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -31.42 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.99 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2717859 120 - 27905053 CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCGAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((.(((((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... ( -34.00) >DroSec_CAF1 28133 120 - 1 CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((.(((((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... ( -34.30) >DroSim_CAF1 29321 120 - 1 CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((.(((((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... ( -34.30) >DroEre_CAF1 29211 120 - 1 CUGUUGGCCAUAAGAAAGGCCAGCGACGAAAAGCGGUAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((((.(((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... ( -30.10) >DroYak_CAF1 28930 120 - 1 CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((.(((((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... ( -34.30) >DroAna_CAF1 28034 120 - 1 CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCUGGAAAACGAGAUACUCGACUCACCAGAAGUCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((.....((((....((((...)))).....))))...............((((..(((........)))..))))...........))))... ( -28.50) >consensus CUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGAGCUGCAGAUGUAAGUAAAUCAAAUAAAAUCACGUGUCAUU ..(((((((........)))))))((((....((.(((((.((((...))))..)))))...((((((........))).))))).(((........))).............))))... (-31.42 = -31.50 + 0.08)

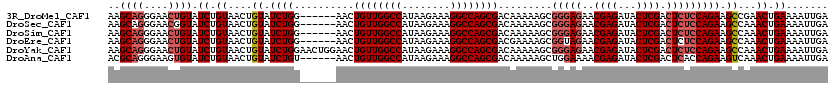
| Location | 2,717,899 – 2,718,013 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2717899 114 - 27905053 AAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCGAACUGAAAAUUGA ..((((....)))).((.((....((.(((((------(..((((((((........))))))))...........(((..((((...)))).))))))))).))...)).))....... ( -33.20) >DroSec_CAF1 28173 114 - 1 AAGCAGGGAACGGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGA ...(((((((((((.......))))).)))((------..(((((((((........)))))))...........(((((.((((...))))..)))))))...))...)))........ ( -32.10) >DroSim_CAF1 29361 114 - 1 AAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGA ..((((....)))).((.((....((.(((((------(..((((((((........))))))))...........(((..((((...)))).))))))))).))...)).))....... ( -33.20) >DroEre_CAF1 29251 114 - 1 AAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACGAAAAGCGGUAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGA ..((((....)))).((.((....((.(((((------(..((((((((........))))))))................((((...))))....)))))).))...)).))....... ( -31.50) >DroYak_CAF1 28970 120 - 1 AAGCAGGGAACUGUAUCUGUAACUGUAUCUGGAACUGGAACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGA ..((((....))))...............(((..(((((..((((((((........))))))))...........(((..((((...)))).))))))))...)))............. ( -33.20) >DroAna_CAF1 28074 114 - 1 ACGCAGGGAAGUGUAUCUGUAACUGUAUCUGU------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCUGGAAAACGAGAUACUCGACUCACCAGAAGUCAAACUGAAAAUUGA ..(((((..(((.........)))...)))))------....(((((((........)))))))(((......((((....((((...)))).....))))..))).............. ( -28.50) >consensus AAGCAGGGAACUGUAUCUGUAACUGUAUCUGG______AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGAACGAGAUACUCGACUCUCCAGAAGCCAAACUGAAAAUUGA ..((((....)))).((.((....((.((((..........((((((((........)))))))).........(((((..((((...)))).))))))))).))...)).))....... (-26.29 = -27.15 + 0.86)
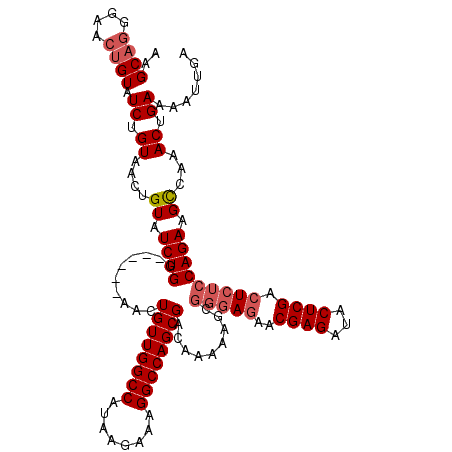
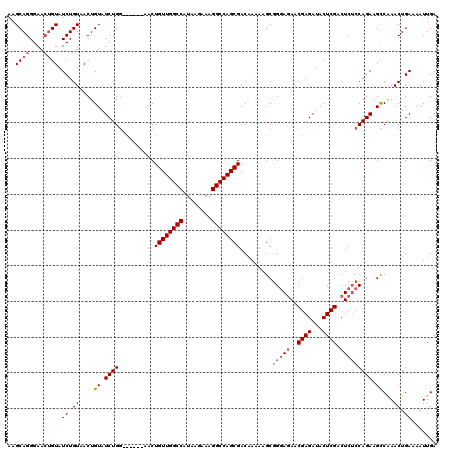
| Location | 2,717,939 – 2,718,053 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2717939 114 - 27905053 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGCCCGAAUGAAGAAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGA ((((.((.(((((....))))).)).))))(((.....(((.((((....)))).)))..............------...((((((((........))))))))........))).... ( -32.80) >DroSec_CAF1 28213 114 - 1 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGCCCGAAUGAAGAAGCAGGGAACGGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGA ((((.((.(((((....))))).)).))))(((.....(((.((((...((((...))))..)))).)))..------...((((((((........))))))))........))).... ( -32.60) >DroSim_CAF1 29401 114 - 1 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGCCCGAAUGAAGAAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGA ((((.((.(((((....))))).)).))))(((.....(((.((((....)))).)))..............------...((((((((........))))))))........))).... ( -32.80) >DroEre_CAF1 29291 114 - 1 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGGCCAAAUGAAGAAGCAGGGAACUGUAUCUGUAACUGUAUCUGG------AACUGUUGGCCAUAAGAAAGGCCAGCGACGAAAAGCGGUAGA (((.....(((.....(((((((((.(((.........(((.((((....)))).)))....))))))))))------)).((((((((........)))))))).)))..)))...... ( -32.52) >DroYak_CAF1 29010 120 - 1 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGGCUGAAUGAAGAAGCAGGGAACUGUAUCUGUAACUGUAUCUGGAACUGGAACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGA (((..((.((...((((((((((((.(((..((.....(((.((((....)))).))).)).)))))))))))).)))....(((((((........)))))))))))...)))...... ( -33.70) >DroAna_CAF1 28114 114 - 1 GCUAAUGAUCGAAUCAUUUCGGAUAAUGGCCGGGCCAAAGACGCAGGGAAGUGUAUCUGUAACUGUAUCUGU------AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCUGGAAA ((((.((.(((((....))))).)).))))..((((((((..(((((..(((.........)))...)))))------..)).))))))..........(((((........)))))... ( -34.20) >consensus GCUAAUGAUCGAAUCAUUUCGGAUAAUGGCCCGAAUGAAGAAGCAGGGAACUGUAUCUGUAACUGUAUCUGG______AACUGUUGGCCAUAAGAAAGGCCAGCGACAAAAAGCGGGAGA .(((.((.(((((....))))).)).)))((((.....(((.((((....)))).))).......................((((((((........))))))))........))))... (-28.24 = -28.30 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:25 2006