
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,636,619 – 2,636,848 |
| Length | 229 |
| Max. P | 0.782261 |

| Location | 2,636,619 – 2,636,728 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -35.49 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2636619 109 + 27905053 ACAC-----A------CAAGGGGUAGCAAGGACUUUGGAAGCAAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGCCAAAAGGCAGCUGGGACACAGUCGCGCUUGCCUCUGUGAUC ...(-----(------((.((((((((.....(((((........)))))............((((.(..((((.(.(((....))).)))))..)....)))))).))))))))))... ( -33.90) >DroSec_CAF1 36935 109 + 1 ACAC-----A------CAGGGGGUAGCAAGGACUUUGGAAGCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCUGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUC ...(-----(------((((((..........(((((........))))).......((((.((((.(..((((.(((.(....).)))))))..)....)))).))))))))))))... ( -40.70) >DroSim_CAF1 25146 109 + 1 ACAC-----A------CAAGGGGUAGCAAGGACUUUGGAAGCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCUGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUC ...(-----(------((.((((((((.....(((((........)))))............((((.(..((((.(((.(....).)))))))..)....)))))).))))))))))... ( -38.30) >DroEre_CAF1 27640 119 + 1 ACACAAUCCACAUGCACAAGGGGUAGCAAGGACU-UGGAAGCGAAUAAAGAGUAAUAUAAGGGUGCAGGACCAGGCAGGCGAAAGGCAGCUGGGACAGAGUCGCAUUUGCCUCUGUGAUC ..............((((.(((((((....((((-(....(((.......(....).......))).(..((((.(.(.(....).).)))))..).)))))....)))))))))))... ( -31.74) >DroYak_CAF1 18585 114 + 1 ACACAAUCCA------CAAGGGGUAGCAAGGACUUUGGAAGCGAGUGAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGCCAAAAGGCAGCUGGGACAAAGUCGCAUUUGCCUCUGUGAUC ........((------((.(((((((....(((((((..(((...................(((.....))).....(((....))).)))....)))))))....)))))))))))... ( -32.80) >consensus ACAC_____A______CAAGGGGUAGCAAGGACUUUGGAAGCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUC ..................((((((((...((((((((..(((...................(((.....))).....(.(....).).)))....))))))).)..))))))))...... (-25.04 = -25.28 + 0.24)

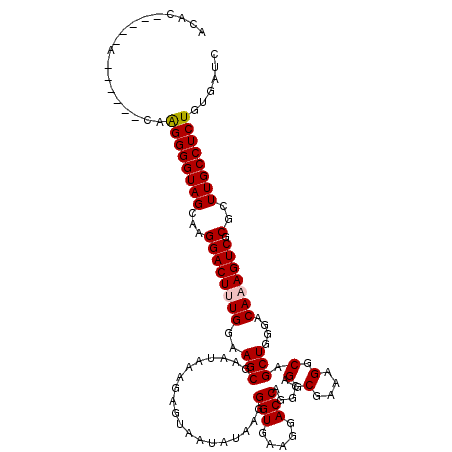
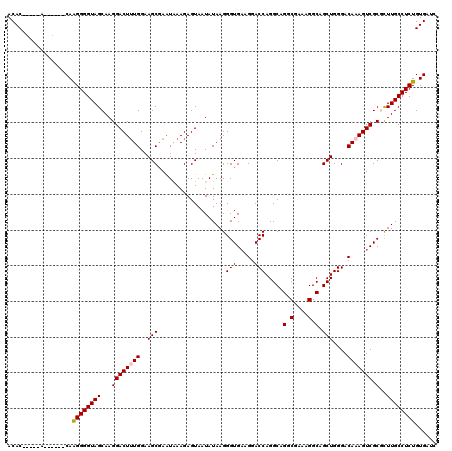
| Location | 2,636,648 – 2,636,768 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -41.04 |
| Consensus MFE | -32.78 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2636648 120 + 27905053 GCAAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGCCAAAAGGCAGCUGGGACACAGUCGCGCUUGCCUCUGUGAUCAUCCAGCUGAAGCUCGCUGCGAAGUGCUCCACUUUUCAGC .........((((.......((((((....((.(((.(((....))).))).)).(((((..((....))..))))).)))))).......))))((((.((((((...)))))).)))) ( -41.64) >DroSec_CAF1 36964 120 + 1 GCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCUGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUCAUCCAGCUGAAGCUCGCUGCGAAGUGCUCCACUUUUCAGC .........((((.......((((((.(..((((.(((.(....).)))))))..)....(((((........))))))))))).......))))((((.((((((...)))))).)))) ( -41.84) >DroSim_CAF1 25175 120 + 1 GCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCUGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUCAUCCAGCUGAAGCUCGCUGCGAAGUGCUCCACUUUUCAGC .........((((.......((((((.(..((((.(((.(....).)))))))..)....(((((........))))))))))).......))))((((.((((((...)))))).)))) ( -41.84) >DroEre_CAF1 27679 120 + 1 GCGAAUAAAGAGUAAUAUAAGGGUGCAGGACCAGGCAGGCGAAAGGCAGCUGGGACAGAGUCGCAUUUGCCUCUGUGAUCAUCCAGCUGAAGCUCGCUGCAAAGUGCUCCACUUUUCAAC ..................(((((((.((.((...((((.(((....((((((((((((((..((....)))))))).....))))))))....)))))))...)).)).))))))).... ( -40.90) >DroYak_CAF1 18619 120 + 1 GCGAGUGAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGCCAAAAGGCAGCUGGGACAAAGUCGCAUUUGCCUCUGUGAUCAUCCAGCCAAUGCUCGCUGCAAAGUGCUCCACUUUUCAGC .........(((((......((((((.(..((((.(.(((....))).)))))..)....(((((........)))))))))))......)))))((((.((((((...)))))).)))) ( -39.00) >consensus GCGAAUAAAGAGUAAUAUAAGGGUGAAGGACCAGGCAGGCGAAAGGCAGCUGGGACAAAGUCGCGCUUGCCUCUGUGAUCAUCCAGCUGAAGCUCGCUGCGAAGUGCUCCACUUUUCAGC .........((((.......((((((.(..((((.(.(.(....).).)))))..)....(((((........))))))))))).......))))((((.((((((...)))))).)))) (-32.78 = -32.70 + -0.08)

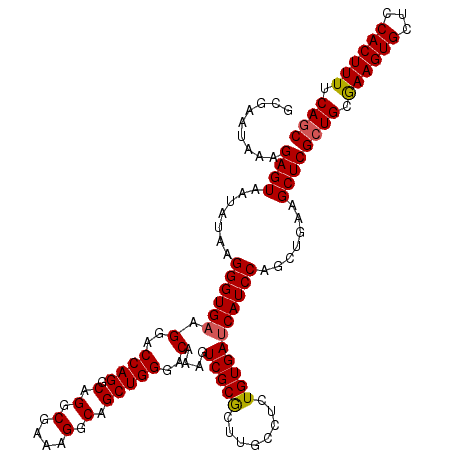
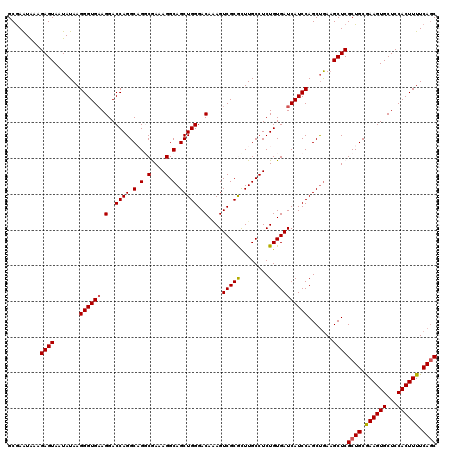
| Location | 2,636,728 – 2,636,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -34.30 |
| Energy contribution | -33.78 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2636728 120 - 27905053 CGUGGGUGCUGUUGGGCUGGGGUGAUGUAAUAUAUUUUGCGGACUUCGAUUUGGCAAGACGGCGAUAAUUUGGCAAAGGUGCUGAAAAGUGGAGCACUUCGCAGCGAGCUUCAGCUGGAU ......(((((((..((((((((..(((((......))))).))))).....)))..)))))))...(((..((..((((((((..(((((...)))))..))))..))))..))..))) ( -37.10) >DroSec_CAF1 37044 120 - 1 CGGGGGUGCUGUUGGGCUGGGGUGAUGUAAUAUUUUUUGCGGACUUCGAUUUGGCAAGACGGCGAUAAUUUGGCAAAGGUGCUGAAAAGUGGAGCACUUCGCAGCGAGCUUCAGCUGGAU ......(((((((..((((((((..(((((......))))).))))).....)))..)))))))...(((..((..((((((((..(((((...)))))..))))..))))..))..))) ( -37.10) >DroSim_CAF1 25255 120 - 1 CGGGGGUGCUGUUGGGCUGGGGUGAUGUAAUAUUUUUUGCGGUCUUCGAUUUGGCAAGACGGCGAUAAUUUGGCAAAGGUGCUGAAAAGUGGAGCACUUCGCAGCGAGCUUCAGCUGGAU ..........(((.(((((((((..(((...(((..((((.((((((......).))))).)))).)))...)))....(((((..(((((...)))))..))))).))))))))).))) ( -38.60) >DroEre_CAF1 27759 120 - 1 CGGGGGUGCCGUUGCGCUGGGGUGAUGUAAUAUAUUUUGCGAAUUUCGAUUUGGCAAGACGGCGAUAAUUUGGCAAAGGUGUUGAAAAGUGGAGCACUUUGCAGCGAGCUUCAGCUGGAU ......(((((((..((..((.((((((((......)))))....))).))..))..)))))))...(((..((..((((((((.((((((...)))))).))))..))))..))..))) ( -35.60) >DroYak_CAF1 18699 120 - 1 CGGGGUUGCUGAUGGGUUGAAAUGAUGUAAUAUAUUUUGCUGAUUUUGAUUUGGCAAGACGGCAAUAAUUUGGCAAAGGUGCUGAAAAGUGGAGCACUUUGCAGCGAGCAUUGGCUGGAU ....(((((((.((.(((.....))).)).....((((((..(.......)..))))))))))))).(((..((.....(((((.((((((...)))))).))))).......))..))) ( -33.30) >consensus CGGGGGUGCUGUUGGGCUGGGGUGAUGUAAUAUAUUUUGCGGACUUCGAUUUGGCAAGACGGCGAUAAUUUGGCAAAGGUGCUGAAAAGUGGAGCACUUCGCAGCGAGCUUCAGCUGGAU ......(((((((..((..((.((((((((......)))))....))).))..))..)))))))...(((..((..((((((((..(((((...)))))..))))..))))..))..))) (-34.30 = -33.78 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:00 2006