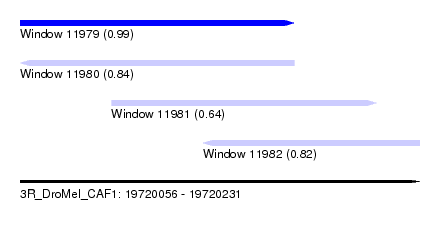
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 19,720,056 – 19,720,231 |
| Length | 175 |
| Max. P | 0.994255 |

| Location | 19,720,056 – 19,720,176 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
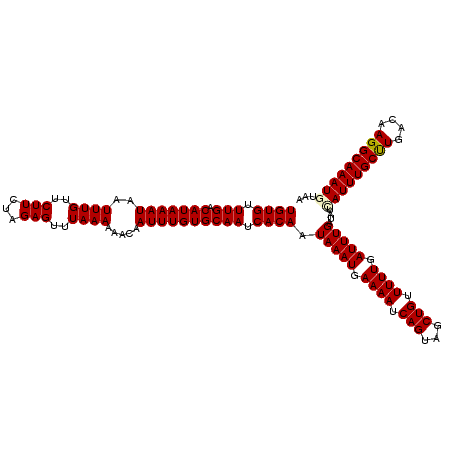
>3R_DroMel_CAF1 19720056 120 + 27905053 UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCUUGACAAGGCAAAUGUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).)))))....(((((((((....)))))))))... ( -25.40) >DroSec_CAF1 33661 120 + 1 UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCCUGACAAGGCAAAUUUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).))))).....((((((((....)))))))).... ( -25.20) >DroSim_CAF1 39037 120 + 1 UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCCUGACAAGGCAAAUUUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).))))).....((((((((....)))))))).... ( -25.20) >DroEre_CAF1 34542 120 + 1 UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGCUUUUGAUUUGAUAUUAUUUGCUUGACAAGGCAAAUGUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).)))))....(((((((((....)))))))))... ( -23.80) >DroYak_CAF1 34497 120 + 1 UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGCUUUUGAUUUGAUAUUAUUUGCUUGACAAGGCAAAUGUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).)))))....(((((((((....)))))))))... ( -23.80) >consensus UGUGUUUGACAUAAAUAAUUUGUUCUUCUAGAGUUUAAAAAACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCUUGACAAGGCAAAUGUAA ((((.(((.(((((((..((((..(((...)))..)))).....)))))))))).)))).(((((.((((.(((...))).)))).)))))....(((((((((....)))))))))... (-24.44 = -24.52 + 0.08)

| Location | 19,720,056 – 19,720,176 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19720056 120 - 27905053 UUACAUUUGCCUUGUCAAGCAAAUGAUAUCAAAUCAAAAACAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ...(((((((........))))))).....((((.((((.(((...))).)))).))))..((((.(((((.((((((((((((........))))))))....)))).)).))).)))) ( -19.80) >DroSec_CAF1 33661 120 - 1 UUAAAUUUGCCUUGUCAGGCAAAUGAUAUCAAAUCAAAAACAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ....((((((((....))))))))(((((.((((.((((.(((...))).)))).)))).(((((....))))).(((((((((........))))))))).......)))))....... ( -22.30) >DroSim_CAF1 39037 120 - 1 UUAAAUUUGCCUUGUCAGGCAAAUGAUAUCAAAUCAAAAACAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ....((((((((....))))))))(((((.((((.((((.(((...))).)))).)))).(((((....))))).(((((((((........))))))))).......)))))....... ( -22.30) >DroEre_CAF1 34542 120 - 1 UUACAUUUGCCUUGUCAAGCAAAUAAUAUCAAAUCAAAAGCAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ....((((((........))))))......((((.((((.(((...))).)))).))))..((((.(((((.((((((((((((........))))))))....)))).)).))).)))) ( -16.60) >DroYak_CAF1 34497 120 - 1 UUACAUUUGCCUUGUCAAGCAAAUAAUAUCAAAUCAAAAGCAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ....((((((........))))))......((((.((((.(((...))).)))).))))..((((.(((((.((((((((((((........))))))))....)))).)).))).)))) ( -16.60) >consensus UUACAUUUGCCUUGUCAAGCAAAUGAUAUCAAAUCAAAAACAGCUACUGAUUUUCAUUUAUUGUGAUUGCACAAAUUGUUUUUUAAACUCUAGAAGAACAAAUUAUUUAUGUCAAACACA ....((((((........))))))......((((.((((.(((...))).)))).))))..((((.(((((.((((((((((((........))))))))....)))).)).))).)))) (-17.36 = -17.36 + -0.00)

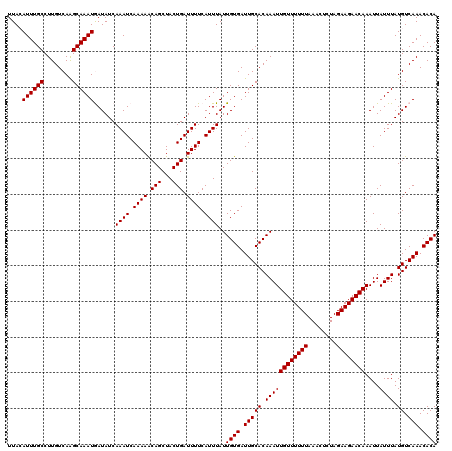
| Location | 19,720,096 – 19,720,212 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.90 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
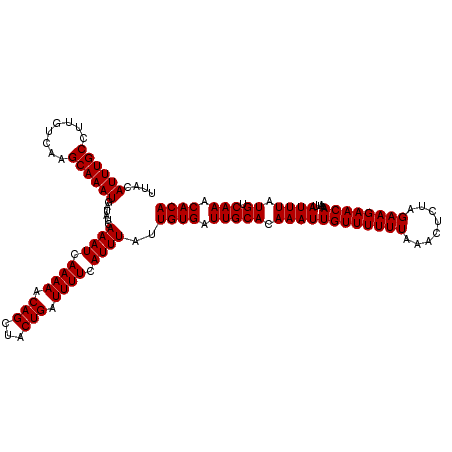
>3R_DroMel_CAF1 19720096 116 + 27905053 AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCUUGACAAGGCAAAUGUAAUUGUU----GUGGAUGUUCUUUAAACUUAGUGGCUAAGUA ........(.(((.(((((((((....(((((((.........))))))).....(((((((((....)))))))))...)))))----)))).))).).....((((((...)))))). ( -27.40) >DroSec_CAF1 33701 120 + 1 AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCCUGACAAGGCAAAUUUAAUUGUUGUUUGUGGAUGUUCUUUAAACUUAGUGGCUAAGUA ........(.(((.(((((((((((.((((.(((...))).)))).))))).....((((((((....))))))))...........)))))).))).).....((((((...)))))). ( -25.50) >DroSim_CAF1 39077 120 + 1 AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCCUGACAAGGCAAAUUUAAUUGUUGUUUGUGGAUGUUCUUUAAACUUAGUGGCUAAGUA ........(.(((.(((((((((((.((((.(((...))).)))).))))).....((((((((....))))))))...........)))))).))).).....((((((...)))))). ( -25.50) >DroEre_CAF1 34582 99 + 1 AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGCUUUUGAUUUGAUAUUAUUUGCUUGACAAGGCAAAUGUAAUUGUU----GUGGAUGUUCUUUA----------------- ........(.(((.(((((((((....(((((((.........))))))).....(((((((((....)))))))))...)))))----)))).))).)....----------------- ( -22.50) >DroYak_CAF1 34537 116 + 1 AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGCUUUUGAUUUGAUAUUAUUUGCUUGACAAGGCAAAUGUAAUUGUU----UUGGAUGUUCUUUAAACUUAGUGGCUAAGUA ......(((((....)))))..............((((..((((..(..((((..(((((((((....)))))))))..))))..----)..)).(((.....)))..))..)))).... ( -21.60) >consensus AACAAUUUGUGCAAUCACAAUAAAUGAAAAUCAGUAGCUGUUUUUGAUUUGAUAUCAUUUGCUUGACAAGGCAAAUGUAAUUGUU____GUGGAUGUUCUUUAAACUUAGUGGCUAAGUA ........(.(((.((((..(((((.((((.(((...))).)))).)))))....(((((((((....)))))))))............)))).))).).....((((((...)))))). (-19.74 = -20.32 + 0.58)

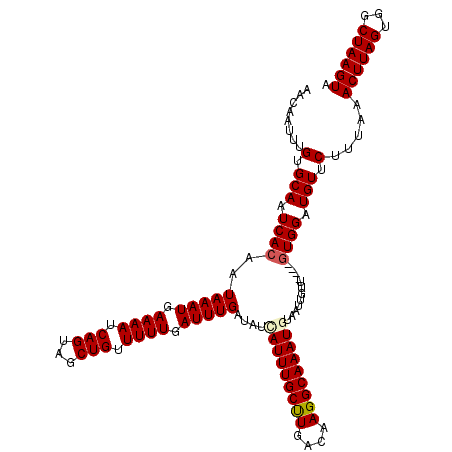
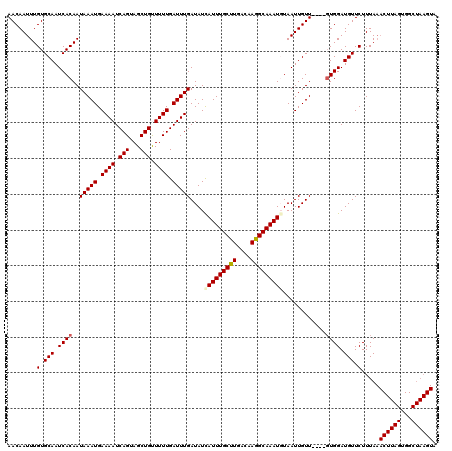
| Location | 19,720,136 – 19,720,231 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -13.75 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 19720136 95 - 27905053 AAUAGGUAACCUAAGCUGAUACUUAGCCACUAAGUUUAAAGAACAUCCAC----AACAAUUACAUUUGCCUUGUCAAGCAAAUGAUAUCAAAUCAAAAA ..((((...))))...((((((((((...)))))................----........(((((((........))))))).)))))......... ( -13.90) >DroSec_CAF1 33741 99 - 1 AAUAGGUAACCUAAGCUGAUACUUAGCCACUAAGUUUAAAGAACAUCCACAAACAACAAUUAAAUUUGCCUUGUCAGGCAAAUGAUAUCAAAUCAAAAA ..((((...))))...((((((((((...))))).............................((((((((....))))))))..)))))......... ( -15.20) >DroSim_CAF1 39117 99 - 1 AAUAGGUAACCUAAGCUGAUACUUAGCCACUAAGUUUAAAGAACAUCCACAAACAACAAUUAAAUUUGCCUUGUCAGGCAAAUGAUAUCAAAUCAAAAA ..((((...))))...((((((((((...))))).............................((((((((....))))))))..)))))......... ( -15.20) >DroYak_CAF1 34577 95 - 1 AAUAGGUAACCUAAGCUGAUACUUAGCCACUAAGUUUAAAGAACAUCCAA----AACAAUUACAUUUGCCUUGUCAAGCAAAUAAUAUCAAAUCAAAAG ..((((...))))...((((((((((...)))))................----.........((((((........))))))..)))))......... ( -10.70) >consensus AAUAGGUAACCUAAGCUGAUACUUAGCCACUAAGUUUAAAGAACAUCCAC____AACAAUUAAAUUUGCCUUGUCAAGCAAAUGAUAUCAAAUCAAAAA ..((((...))))...((((((((((...))))).............................((((((((....))))))))..)))))......... (-12.00 = -12.50 + 0.50)

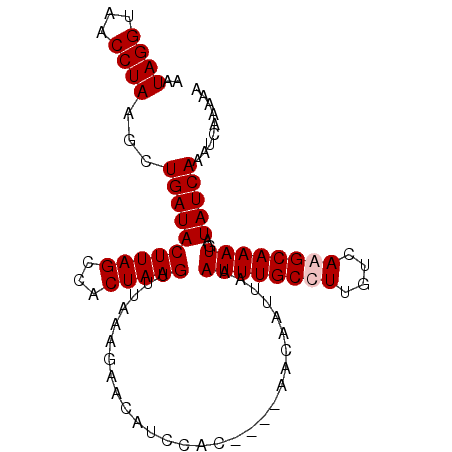
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:19 2006