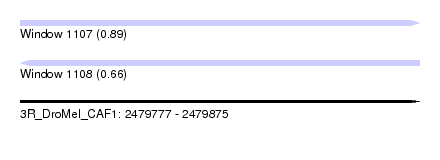
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,479,777 – 2,479,875 |
| Length | 98 |
| Max. P | 0.889916 |

| Location | 2,479,777 – 2,479,875 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
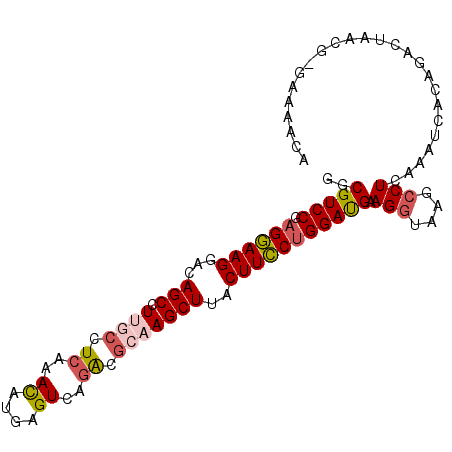
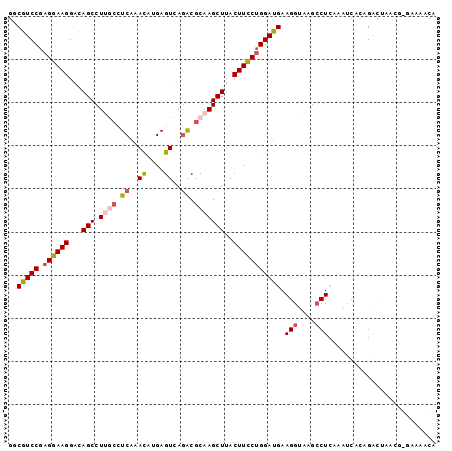
>3R_DroMel_CAF1 2479777 98 + 27905053 GGCGUCCGAGGAAGGACAGCCUUGCCUCAAACAUGAGUCACACGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCUAAUCGCCGACUAAUU-GAAACUA (((((((.((((((...(((.((((((((....))))......)))))))..))))))))))..(((....)))......)))........-....... ( -29.60) >DroVir_CAF1 39085 95 + 1 GCCGUCCCCGAAAGGACAGCCUUGGCUCUAAUAUGAGUCAGACGCAAGCUUACUUUCUGGACGAAGGUAAGACUUAUA-CGGAGUU-UA--AGAAACUA ((.((((......)))).))(((((((((..((((((((.(.(....))((((((((.....))))))))))))))))-.)))).)-))--))...... ( -30.60) >DroPse_CAF1 7034 98 + 1 GUCGUCCGAGGAAGGGUAGCCUAACAUCAAACAAGAGUCAGGCGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCAAACUACAGACUAACG-GAUUACA ((.((((((((((((((.((((.((.((......)))).))))....)))..))))))(..(((.((....)))))..)..........))-))).)). ( -28.40) >DroYak_CAF1 7340 98 + 1 GGCGUCCGAGGAAGGACAGCCUUGCCUCAAACAUGAGUCAGACGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCUAGUCACCGACUAAUU-GGAACUA ..(((((.((((((...(((.((((.((..((....))..)).)))))))..)))))))))))..(((...((..(((((...)))))...-)).))). ( -30.80) >DroAna_CAF1 7338 98 + 1 GGCGUCCGAGGAAGGACAGCCUUGCUUCAAACAUGAGUCAGACGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCUAAUCACCGACAAACA-AACCACA (((((((.((((((...(((.((((.((..((....))..)).)))))))..))))))))))).(((....))).......))........-....... ( -28.20) >DroPer_CAF1 7031 98 + 1 GUCGUCCGAGGAAGGGUAGCCUAACAUCAAACAAGAGUCAGGCGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCAAACUACAGACUAACG-GAUUACA ((.((((((((((((((.((((.((.((......)))).))))....)))..))))))(..(((.((....)))))..)..........))-))).)). ( -28.40) >consensus GGCGUCCGAGGAAGGACAGCCUUGCCUCAAACAUGAGUCAGACGCAAGCUUACUUCCUGGAUGAAGGUAAGCCUCAAAUCACAGACUAACG_GAAAACA ..(((((.((((((...(((.((((.((..((....))..)).)))))))..))))))))))).(((....)))......................... (-20.67 = -21.23 + 0.56)

| Location | 2,479,777 – 2,479,875 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -19.80 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
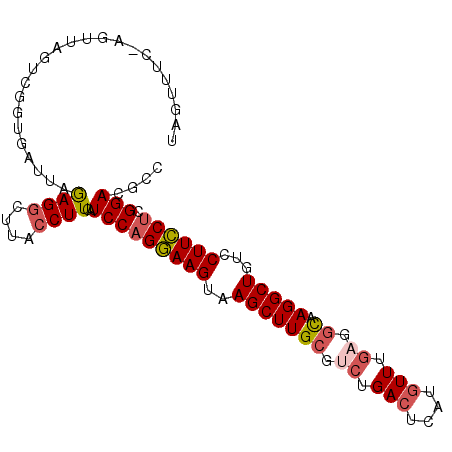
>3R_DroMel_CAF1 2479777 98 - 27905053 UAGUUUC-AAUUAGUCGGCGAUUAGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGUGUGACUCAUGUUUGAGGCAAGGCUGUCCUUCCUCGGACGCC ..(((((-((..((((((((.....((((((((.(((......))))))))))).))).))))).....)))))))..(((.((((......))))))) ( -32.20) >DroVir_CAF1 39085 95 - 1 UAGUUUCU--UA-AACUCCG-UAUAAGUCUUACCUUCGUCCAGAAAGUAAGCUUGCGUCUGACUCAUAUUAGAGCCAAGGCUGUCCUUUCGGGGACGGC ........--..-.....((-((..((.(((((.(((.....))).)))))))))))..((.(((......))).))..((((((((....)))))))) ( -24.60) >DroPse_CAF1 7034 98 - 1 UGUAAUC-CGUUAGUCUGUAGUUUGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGCCUGACUCUUGUUUGAUGUUAGGCUACCCUUCCUCGGACGAC .......-.....(((((.((....((((((((.(((......)))))))))))..(((((((((......)).)))))))........)))))))... ( -29.90) >DroYak_CAF1 7340 98 - 1 UAGUUCC-AAUUAGUCGGUGACUAGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGUCUGACUCAUGUUUGAGGCAAGGCUGUCCUUCCUCGGACGCC (((((((-........)).))))).((((((((.(((......)))))))))))(((((((((....))).((((.((((....)))))))))))))). ( -33.10) >DroAna_CAF1 7338 98 - 1 UGUGGUU-UGUUUGUCGGUGAUUAGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGUCUGACUCAUGUUUGAAGCAAGGCUGUCCUUCCUCGGACGCC .......-........((((....((((....))))..(((((((((..(((((((.((.(((....))).)).)))).)))...)))))).))))))) ( -30.10) >DroPer_CAF1 7031 98 - 1 UGUAAUC-CGUUAGUCUGUAGUUUGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGCCUGACUCUUGUUUGAUGUUAGGCUACCCUUCCUCGGACGAC .......-.....(((((.((....((((((((.(((......)))))))))))..(((((((((......)).)))))))........)))))))... ( -29.90) >consensus UAGUUUC_AGUUAGUCGGUGAUUAGAGGCUUACCUUCAUCCAGGAAGUAAGCUUGCGUCUGACUCAUGUUUGAGGCAAGGCUGUCCUUCCUCGGACGCC ........................((((....))))..(((((((((..(((((((.((.(((....))).)).)).)))))...)))))).))).... (-19.80 = -20.33 + 0.53)

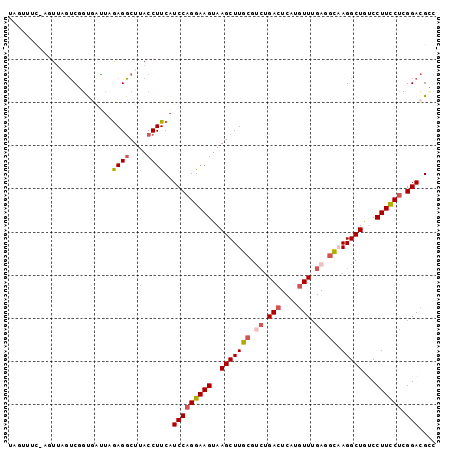
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:40 2006