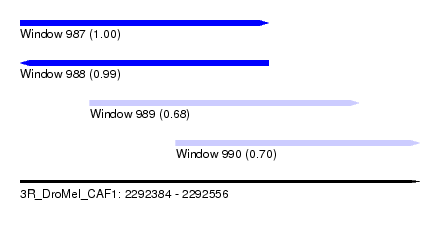
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,292,384 – 2,292,556 |
| Length | 172 |
| Max. P | 0.997503 |

| Location | 2,292,384 – 2,292,491 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292384 107 + 27905053 UAAGCCGCGUUUG---------CCCAAAAACGGUCAACA-GACCACAUUCAACGACCACUUGGC--CCAUGUCGC-UGGCAGGACCGCCAAUGUAAUAAUAGCAAAGGCAAACAGCAGCG ...((.(((((((---------((.......((((....-)))).............(((((((--((.(((...-..)))))...))))).))............))))))).)).)). ( -31.70) >DroSec_CAF1 28642 107 + 1 UAAGCCGCGUUUG---------CCCAAAAACGGUCAAAC-GACCACAUUCAACGACCACUUGGU--CCAUGUCGC-UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCG ...((.(((((((---------((.......((((....-)))).................(((--((.(((...-..))))))))....................))))))).)).)). ( -33.80) >DroSim_CAF1 30268 108 + 1 UAAGCCGCGUUUG---------CCCAAAAACGGUCAAACUUACCACAUUCAACGACCACUUGGU--CCAUGUCGC-UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCG ...((.(((((((---------((.......((((..................))))....(((--((.(((...-..))))))))....................))))))).)).)). ( -32.27) >DroYak_CAF1 27447 96 + 1 UAAGCUGCGUUUGGCCGUUUUGACCAAAAACGGUCAACC-GACCACAUUCAACGACCACUUGG-----------------------GCCAAUAUAAUAAUAGCAAAAGCAAACUGCAGCG ...(((((..(((((((((((.....)))))((((....-))))..................)-----------------------)))))..........((....)).....))))). ( -30.10) >DroAna_CAF1 28219 103 + 1 UAAGCCGCGUUUCA--------CCCAAAAACGGUCAAAU-GACCACAUUCAUCGGCCGCUUGGUGUCCACGGACCACGUUAUCCCAGCCAAUAUAAUAAUAACAAAGGCAAA-------- (((((.((((((..--------.....))))((((....-))))..........)).)))))(((((....)).))).........(((.................)))...-------- ( -21.33) >consensus UAAGCCGCGUUUG_________CCCAAAAACGGUCAAAC_GACCACAUUCAACGACCACUUGGU__CCAUGUCGC_UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCG ...(((..(((....................((((.....))))...............(((((.....((((....)))).....))))).........)))...)))........... (-14.60 = -14.64 + 0.04)

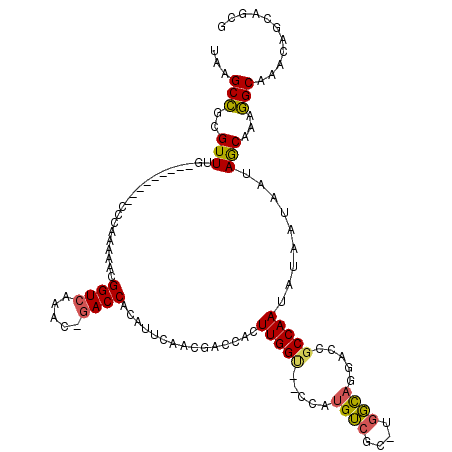
| Location | 2,292,384 – 2,292,491 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292384 107 - 27905053 CGCUGCUGUUUGCCUUUGCUAUUAUUACAUUGGCGGUCCUGCCA-GCGACAUGG--GCCAAGUGGUCGUUGAAUGUGGUC-UGUUGACCGUUUUUGGG---------CAAACGCGGCUUA .(((((.((((((((..((.......(((((((((....)))).-.((((...(--(((....)))))))))))))((((-....))))))....)))---------))))))))))... ( -42.20) >DroSec_CAF1 28642 107 - 1 CGCUGCUGUUUGCCUUUGCUAUUAUUAUAUUGGCGGUCCUGCCA-GCGACAUGG--ACCAAGUGGUCGUUGAAUGUGGUC-GUUUGACCGUUUUUGGG---------CAAACGCGGCUUA .(((((.((((((((..((((.........))))((((..((..-((.((((.(--(((....)))).....)))).)).-))..))))......)))---------))))))))))... ( -41.00) >DroSim_CAF1 30268 108 - 1 CGCUGCUGUUUGCCUUUGCUAUUAUUAUAUUGGCGGUCCUGCCA-GCGACAUGG--ACCAAGUGGUCGUUGAAUGUGGUAAGUUUGACCGUUUUUGGG---------CAAACGCGGCUUA .(((((.((((((((..((((.........))))((((..((..-((.((((.(--(((....)))).....)))).))..))..))))......)))---------))))))))))... ( -42.10) >DroYak_CAF1 27447 96 - 1 CGCUGCAGUUUGCUUUUGCUAUUAUUAUAUUGGC-----------------------CCAAGUGGUCGUUGAAUGUGGUC-GGUUGACCGUUUUUGGUCAAAACGGCCAAACGCAGCUUA .(((((.(.(..(((..((((.........))))-----------------------..)))..).)........(((((-(.(((((((....)))))))..))))))...)))))... ( -34.30) >DroAna_CAF1 28219 103 - 1 --------UUUGCCUUUGUUAUUAUUAUAUUGGCUGGGAUAACGUGGUCCGUGGACACCAAGCGGCCGAUGAAUGUGGUC-AUUUGACCGUUUUUGGG--------UGAAACGCGGCUUA --------...(((.............(((((((((((((......)))).(((...)))..)))))))))...((((((-....))))(((((....--------.))))))))))... ( -29.90) >consensus CGCUGCUGUUUGCCUUUGCUAUUAUUAUAUUGGCGGUCCUGCCA_GCGACAUGG__ACCAAGUGGUCGUUGAAUGUGGUC_GUUUGACCGUUUUUGGG_________CAAACGCGGCUUA .(((((.(((((((..((.((....))))..))).......................(((((((((((................))))))..)))))...........)))))))))... (-18.15 = -18.95 + 0.80)

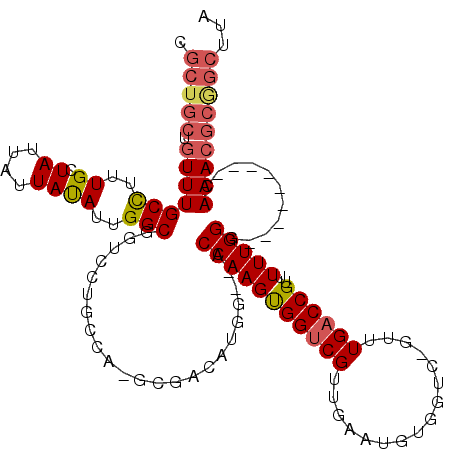
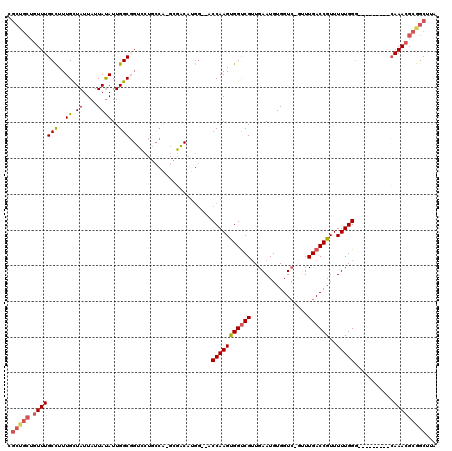
| Location | 2,292,414 – 2,292,530 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292414 116 + 27905053 GACCACAUUCAACGACCACUUGGC--CCAUGUCGC-UGGCAGGACCGCCAAUGUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCU- ................((((((((--(((((((((-((.(.(....(((..(((.......)))..)))...).)))))))))).((((((.....)))))).....))))))))))..- ( -44.70) >DroSec_CAF1 28672 116 + 1 GACCACAUUCAACGACCACUUGGU--CCAUGUCGC-UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCU- ((((.((((....((((....)))--)((((((((-((((......)))............((....)).......)))))))))((((((.....))))))))))..)))).......- ( -40.90) >DroSim_CAF1 30299 116 + 1 UACCACAUUCAACGACCACUUGGU--CCAUGUCGC-UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCU- ................((((((..--(((((((((-((((......)))............((....)).......)))))))).((((((.....)))))).....))..))))))..- ( -40.10) >DroYak_CAF1 27486 96 + 1 GACCACAUUCAACGACCACUUGG-----------------------GCCAAUAUAAUAAUAGCAAAAGCAAACUGCAGCGAUAUGGGAUCCGAUUUGAAUCCAAUGCGGGUCGAGUGCU- ................(((((((-----------------------.((..................((.....)).(((...((((.((......)).)))).))))).)))))))..- ( -20.10) >DroAna_CAF1 28250 106 + 1 GACCACAUUCAUCGGCCGCUUGGUGUCCACGGACCACGUUAUCCCAGCCAAUAUAAUAAUAACAAAGGCAAA---------UAUGGGAUCCGAUUUGCAC-----UUGGGUCGAGUGCCA ...........((((((.(..(((((...((((........((((((((.................)))...---------..)))))))))....))))-----).)))))))...... ( -25.83) >consensus GACCACAUUCAACGACCACUUGGU__CCAUGUCGC_UGGCAGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCU_ .....(((((.....(((.(((((.....((((....)))).....)))))..........((....))..............)))((((((..(((....)))..)))))))))))... (-20.90 = -21.70 + 0.80)

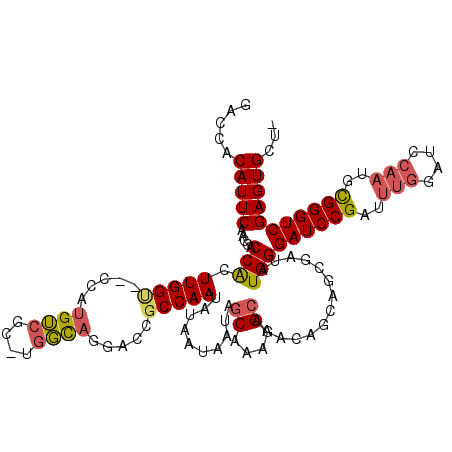
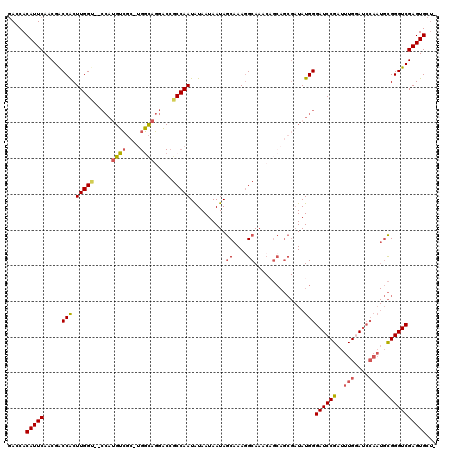
| Location | 2,292,451 – 2,292,556 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2292451 105 + 27905053 AGGACCGCCAAUGUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCUGGUGCAUCCGCAUCCAUAUGUGGACU .(.((((((..(((.......)))..)))....((((.((((...((((((.....)))))).......))))..))))))).).(((((((....))))))).. ( -36.60) >DroSec_CAF1 28709 105 + 1 AGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCUGGUCCAUCCGCAUCCAUAUGUGGACU .((((((((.................)))....((((.((((...((((((.....)))))).......))))..))))))))).(((((((....))))))).. ( -38.53) >DroSim_CAF1 30336 105 + 1 AGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCUGGUGCAUCCGCAUCCAUACCUGGACU ....((((.............((....((.....)).))......((((((.....))))))...))))(((.((...((((((....))).)))...)).))). ( -30.60) >DroYak_CAF1 27509 99 + 1 ------GCCAAUAUAAUAAUAGCAAAAGCAAACUGCAGCGAUAUGGGAUCCGAUUUGAAUCCAAUGCGGGUCGAGUGCUGGUGCAUCCGUAUCCAUAUGUGGCAU ------((((.((((............((.....)).(.(((((((((((((..(((....)))..))))))..((((....)))))))))))).)))))))).. ( -31.40) >consensus AGGACCGCCAAUAUAAUAAUAGCAAAGGCAAACAGCAGCGAUAUGGGAUCCGAUUUGGAUCCAAUGCGGGUCGAGUGCUGGUGCAUCCGCAUCCAUAUGUGGACU .......(((...........((....((.....)).)).((((((((((((..(((....)))..))))))..((((.((.....)))))))))))).)))... (-25.54 = -25.60 + 0.06)

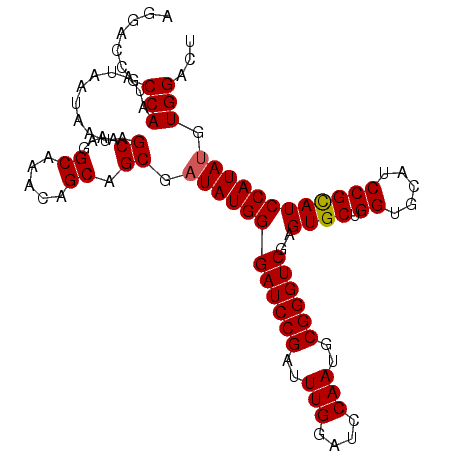

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:42 2006