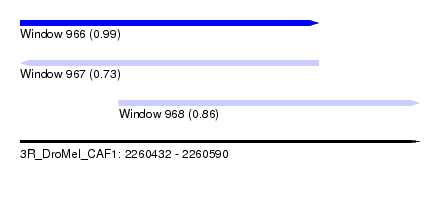
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,260,432 – 2,260,590 |
| Length | 158 |
| Max. P | 0.990133 |

| Location | 2,260,432 – 2,260,550 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
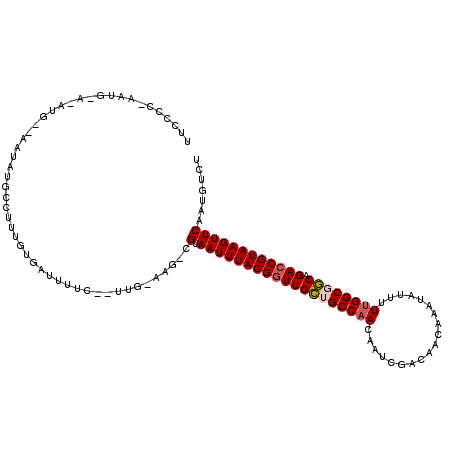
>3R_DroMel_CAF1 2260432 118 + 27905053 UUCCCCGAAUGAAAAUGGAAAUAUGUCUUUGUGAUUUUCGAUUGCAAGCCGUAAUUUACGAUUGCUGCCCACAAUCGACAACAAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCU ..........((...((((..((((((((((..(((...)))..)))(((((......((((((.......))))))....((((....)))))))))))).))))..))))...)). ( -27.00) >DroSec_CAF1 51346 105 + 1 UUCCCCUAAUGAAUAUG--AAUAUGCUUUUGUGAUUUUC-----------GUAAUUUACGGUUGCUGCCAACAAUCGACAACGAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCU .........(((.((((--((.((((....)).)).)))-----------)))((((((((((((((((((.(((((....)))...)).))))))))).))))))))))))...... ( -30.00) >DroSim_CAF1 49497 104 + 1 UUCCCCUAAUGGAAAUG--AAUAUGCUUUUGUGAUUUUC-----------GUAAUUUACGGUUGCUGCCAACAAUCGACAACGAAUAUUUUUGGCGGC-GACCGUAAGUUCAAUGUCU ........((((((((.--.(((......))).))))))-----------))(((((((((((((((((((.(((((....)))...)).))))))))-)))))))))))........ ( -34.40) >DroEre_CAF1 49699 102 + 1 --------------AUG--AGUUGGCCUUUGUGAUUUUCGGUUGCAAGUCGUAAUUUACGGUUGCAGCCAACAAUCGACAACAAAUAACUUUGGCAGCAGACCGUAAGUUCAAUGUCU --------------..(--(((((....((((((((...((((((((.(((((...)))))))))))))...)))).))))....)))))).((((...((.(....).))..)))). ( -28.90) >DroYak_CAF1 46543 100 + 1 --------------CUC--A-UUUGCCUUUUUGAUUUUCAGUUGUAAGUCGUAAUUUACGGUUGUAGCCAACAAUCGACAACAAAUG-UUUUGGCGACAGACCGUAAGUUCAAUGUCU --------------...--(-(((((....(((.....)))..)))))).(.(((((((((((((.(((((.....((((.....))-))))))).))).)))))))))))....... ( -23.60) >consensus UUCCCC_AAUG_A_AUG__AAUAUGCCUUUGUGAUUUUC__UUG_AAG_CGUAAUUUACGGUUGCUGCCAACAAUCGACAACAAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCU ..................................................(.(((((((((((((((((((...................)))))))).))))))))))))....... (-20.37 = -20.85 + 0.48)

| Location | 2,260,432 – 2,260,550 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.44 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2260432 118 - 27905053 AGACAUUGAACUUACGGUCUGCCGCCAAAAAUAUUUGUUGUCGAUUGUGGGCAGCAAUCGUAAAUUACGGCUUGCAAUCGAAAAUCACAAAGACAUAUUUCCAUUUUCAUUCGGGGAA ..............(((....))).........(((((..(((((((..(((......((((...)))))))..))))))).....)))))......(..((..........))..). ( -24.30) >DroSec_CAF1 51346 105 - 1 AGACAUUGAACUUACGGUCUGCCGCCAAAAAUAUUCGUUGUCGAUUGUUGGCAGCAACCGUAAAUUAC-----------GAAAAUCACAAAAGCAUAUU--CAUAUUCAUUAGGGGAA .((...((((.(((((((.(((.(((((......(((....)))...))))).)))))))))).....-----------(.....)...........))--))...)).......... ( -23.00) >DroSim_CAF1 49497 104 - 1 AGACAUUGAACUUACGGUC-GCCGCCAAAAAUAUUCGUUGUCGAUUGUUGGCAGCAACCGUAAAUUAC-----------GAAAAUCACAAAAGCAUAUU--CAUUUCCAUUAGGGGAA .....(((((.(((((((.-((.(((((......(((....)))...))))).)).))))))).)).)-----------))..................--..(..((....))..). ( -25.50) >DroEre_CAF1 49699 102 - 1 AGACAUUGAACUUACGGUCUGCUGCCAAAGUUAUUUGUUGUCGAUUGUUGGCUGCAACCGUAAAUUACGACUUGCAACCGAAAAUCACAAAGGCCAACU--CAU-------------- ...............(((((.....((((....))))((((.((((.((((.(((((.((((...))))..))))).)))).)))))))))))))....--...-------------- ( -24.40) >DroYak_CAF1 46543 100 - 1 AGACAUUGAACUUACGGUCUGUCGCCAAAA-CAUUUGUUGUCGAUUGUUGGCUACAACCGUAAAUUACGACUUACAACUGAAAAUCAAAAAGGCAAA-U--GAG-------------- ...((((...((((((((.(((.((((..(-((.(((....))).))))))).))))))))))...............((.....))....)...))-)--)..-------------- ( -20.70) >consensus AGACAUUGAACUUACGGUCUGCCGCCAAAAAUAUUUGUUGUCGAUUGUUGGCAGCAACCGUAAAUUACG_CUU_CAA__GAAAAUCACAAAGGCAUAUU__CAU_U_CAUU_GGGGAA ...........(((((((.(((.(((((......(((....)))...))))).))))))))))....................................................... (-18.76 = -18.44 + -0.32)



| Location | 2,260,471 – 2,260,590 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.48 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -22.99 |
| Energy contribution | -24.31 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
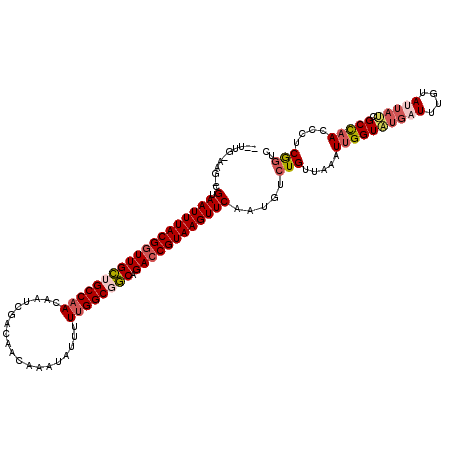
>3R_DroMel_CAF1 2260471 119 + 27905053 GAUUGCAAGCCGUAAUUUACGAUUGCUGCCCACAAUCGACAACAAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCUGUUAAAUUGGUAUGAUUUGUAUUAUCGCCAACCCUCGGUC ........(((((......((((((.......))))))....((((....))))))))).(((((..((........))......((((((((((....))))).)))))....))))) ( -27.50) >DroSec_CAF1 51383 108 + 1 -----------GUAAUUUACGGUUGCUGCCAACAAUCGACAACGAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCUGUUAAAUUGGUAUGUUUUGUAUUAUCGCCAACCCUCGGUC -----------(.(((((((((((((((((((.(((((....)))...)).))))))))).))))))))))).....(((.....((((((((........))).)))))....))).. ( -31.50) >DroSim_CAF1 49534 107 + 1 -----------GUAAUUUACGGUUGCUGCCAACAAUCGACAACGAAUAUUUUUGGCGGC-GACCGUAAGUUCAAUGUCUGUUAAAUUGGUAUGUUUUGUAUUAUCGCCAAUCCUCGAUC -----------(.(((((((((((((((((((.(((((....)))...)).))))))))-))))))))))))............(((((((((........))).))))))........ ( -34.70) >DroEre_CAF1 49722 119 + 1 GGUUGCAAGUCGUAAUUUACGGUUGCAGCCAACAAUCGACAACAAAUAACUUUGGCAGCAGACCGUAAGUUCAAUGUCUGUUAAAUUGGUGUGAUUUGUAUUCCAGCCAACACUCGGUC ((.(((((((((.(((((((((((((.(((((...................))))).))).))))))))))((((.........))))...)))))))))..)).(((.......))). ( -32.01) >DroYak_CAF1 46565 118 + 1 AGUUGUAAGUCGUAAUUUACGGUUGUAGCCAACAAUCGACAACAAAUG-UUUUGGCGACAGACCGUAAGUUCAAUGUCUGUUAAAUUGGUAUGAUUUGUAUAACCGCUUACAAUCAGUC .(((((((((.(((((((((((((((.(((((.....((((.....))-))))))).))).))))))))))..........((((((.....))))))....)).)))))))))..... ( -31.20) >consensus __UUG_AAG_CGUAAUUUACGGUUGCUGCCAACAAUCGACAACAAAUAUUUUUGGCGGCAGACCGUAAGUUCAAUGUCUGUUAAAUUGGUAUGAUUUGUAUUAUCGCCAACCCUCGGUC ...........(.(((((((((((((((((((...................)))))))).)))))))))))).....(((.....((((((((((....))))).)))))....))).. (-22.99 = -24.31 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:20 2006