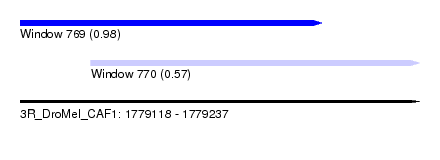
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,779,118 – 1,779,237 |
| Length | 119 |
| Max. P | 0.981929 |

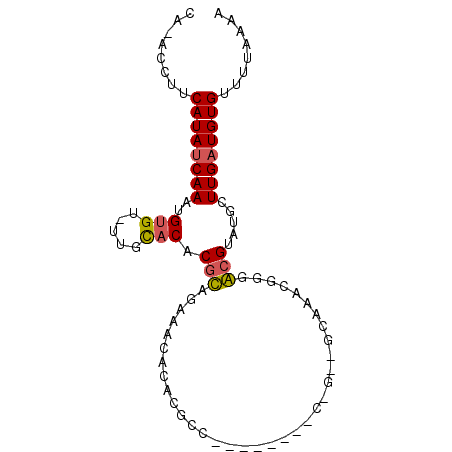
| Location | 1,779,118 – 1,779,208 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -6.57 |
| Energy contribution | -6.43 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.31 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1779118 90 + 27905053 CU-ACCUUCAUAUCAAAUGUGC-UCACACCCGCAGGAGCACAUACCACAUACCACUGCGCCAUACACGGCGUAUACUUGAUGUGUUUUAAAA ..-.....((((((((((((((-((.(.......)))))))))............((((((......))))))...))))))))........ ( -27.40) >DroVir_CAF1 104623 80 + 1 CAAACCUUCAUAUCAAAUGUGUCUUGCACACGUAUAAACA-ACGCC--------C-G--GCGAACGGGACGUAUGCUUGAUGUGUUUUUAAA ........((((((((((((((.....)))))).......-(((((--------(-(--.....)))).)))....))))))))........ ( -22.30) >DroGri_CAF1 90654 83 + 1 CA-ACCUUCAUACCAAAUGUGUCUUGCACACGUAAAAACAAACGCC--------CUGGCGCAAACGGGACGUAUGCUUGAUGUGUUUUUAAA ..-...........(((((..((..(((.((((.........(((.--------...)))........)))).)))..))..)))))..... ( -18.23) >DroYak_CAF1 87758 90 + 1 CU-ACCUUCAUAUCAAAUGUGC-UCACACCCGCCGAAGCACAUGCCACAUACCACUGCGCCAUACACGGCGUAUGCUUGAUGUGUUUUAAAA ..-.....((((((((((((((-((.........).)))))))............((((((......))))))...))))))))........ ( -23.60) >DroMoj_CAF1 96045 79 + 1 CA-ACCUUCAUAUCAAAUGUGUCUUGCACACGUAUAAACA-ACGCC--------U-G--GCAAACGGGACGUAUGCUUGAUGUGUUUUUAAA ..-.....((((((((((((((.....)))))).......-(((((--------(-(--.....)))).)))....))))))))........ ( -20.10) >DroAna_CAF1 81592 78 + 1 CU-ACCUUCAUAUCAAAUGUGU-UGUUCCCCACACUCACACAUCCCGCA------------CUACCGGGCGUAUACUUGAUGUGUUUUAAAA ..-.....((((((((((((((-(((.....)))...))))))((((..------------....)))).......))))))))........ ( -16.50) >consensus CA_ACCUUCAUAUCAAAUGUGU_UUGCACACGCAGAAACACACGCC________C_G__GCAAACGGGACGUAUGCUUGAUGUGUUUUAAAA ........((((((((..(((.....))).(((...................................))).....))))))))........ ( -6.57 = -6.43 + -0.14)


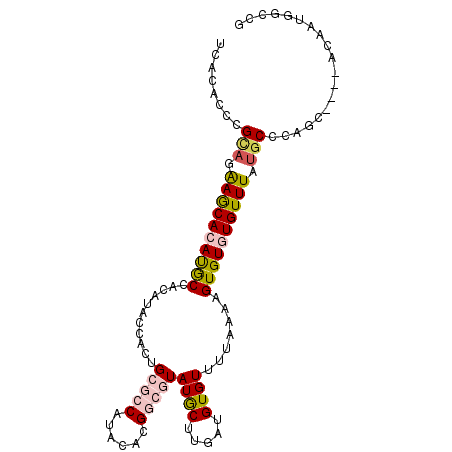
| Location | 1,779,139 – 1,779,237 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1779139 98 + 27905053 UCACACCCGCAGGAGCACAUACCACAUACCACUGCGCCAUACACGGCGUAUACUUGAUGUGUUUUAAAAGUGUGUGUUUAUUCUCAGC----ACAAUGGCCG ........((..(((((((((((((((.....((((((......))))))......)))))........)))))))))).....((..----....)))).. ( -24.90) >DroGri_CAF1 90676 90 + 1 UUGCACACGUAAAAACAAACGCC--------CUGGCGCAAACGGGACGUAUGCUUGAUGUGUUUUUAAAGUGUGUGUUUAUGCGAUGC----ACAAUGGUCG ..((((((.((((((((...((.--------...))(((.(((...))).)))......))))))))..)))))).......((((..----......)))) ( -22.10) >DroSim_CAF1 90548 98 + 1 UCACACCCGCAGGAGCACAUACCACAUACCACUGCGCCAUACACGGCGUAUACUUGAUGUGUUUUAAAAGUGUGUGUUUAUGCCCAGC----ACAAUGGCCG ........(((.(((((((((((((((.....((((((......))))))......)))))........)))))))))).)))(((..----....)))... ( -30.20) >DroEre_CAF1 86579 98 + 1 UCACACCCGCAGAAGCACAUGCCACAUACCACUGCGCCAUACACGGCGUAUGCUUGAUGUGUUUUAAAAGUGUGUGUUUAUGCCCAGC----ACAAUGGCCG ........(((.(((((((((((((((..((.((((((......))))))))....)))))........)))))))))).)))(((..----....)))... ( -29.00) >DroYak_CAF1 87779 102 + 1 UCACACCCGCCGAAGCACAUGCCACAUACCACUGCGCCAUACACGGCGUAUGCUUGAUGUGUUUUAAAAGUGUGUGUUUAUGCCCAGCCAGAACAAUGGCCG ........((..(((((((((((((((..((.((((((......))))))))....)))))........))))))))))..))...((((......)))).. ( -30.30) >DroMoj_CAF1 96067 86 + 1 UUGCACACGUAUAAACA-ACGCC--------U-G--GCAAACGGGACGUAUGCUUGAUGUGUUUUUAAAGUGUGUGUUUAUGCGAUGC----ACAAUGGCCG .((((..((((((((((-(((((--------(-(--.....)))).)))((((((............)))))).)))))))))).)))----)......... ( -25.20) >consensus UCACACCCGCAGAAGCACAUGCCACAUACCACUGCGCCAUACACGGCGUAUGCUUGAUGUGUUUUAAAAGUGUGUGUUUAUGCCCAGC____ACAAUGGCCG ........(((.((((((((((...........(((((......)))))((((.....)))).......)))))))))).)))................... (-17.51 = -17.90 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:07 2006