| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 304,357 – 304,510 |
| Length | 153 |
| Max. P | 0.914275 |

| Location | 304,357 – 304,471 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -23.38 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
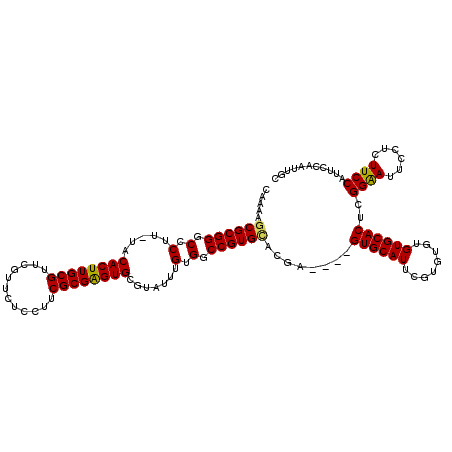
>3R_DroMel_CAF1 304357 114 - 27905053 CGCGAGUGUGUAUUUGUGGCCGUGCACAA----GUGCAUUUGUGUGUGUGCACUCGGGAUUCCUCUUCCAUUCCAAUCGCCGCUCGACCCCCCC--CCCCCCCUUCCACACCAUACCUCG ..((((.((((...(((((..(((((((.----(..(....)..).))))))).(((((((.............)))).)))............--.........)))))..)))))))) ( -30.12) >DroSec_CAF1 39577 106 - 1 CGCGAGUGCGCAUUUGUGACCGUGCACGA----GUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCUAAUUGC-ACUC-------GC--CGCCGCCUUCUGCACUCUACCUCG ...(((((((((((((((......)))))----))))...((.(((.(((((.(..((((.........))))..).)))-)).)-------))--)).........))))))....... ( -37.10) >DroEre_CAF1 43991 114 - 1 CGCGGGUGCGUAUUUGUGGCCGUGAACGAGUGAGUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCCAAUUGCCGCUCGCCCCCGCUUGCCCCCGCUUCCG------CCCACG .(((((.(((.....((((..((((.((.((((((((((........))))))))((((......)))).........)))).))))..)))).))).))))).....------...... ( -42.30) >DroYak_CAF1 43963 91 - 1 CGCGAGUGCGUAUUUGUGGCCGUGUACGA----GUGCAUUCGUAUGUGUGCACACGGAAUUCCUCUUCCAUUCCAAUUGCCGCUUGCCU-------------------------CUCUCG ((.(((.(.(((...(((((.(((((((.----((((....)))).)))))))..((((......)))).........))))).)))).-------------------------))).)) ( -31.00) >consensus CGCGAGUGCGUAUUUGUGGCCGUGCACGA____GUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCCAAUUGCCGCUCGCCC___CC__CCCCCCCUUCCG______ACCUCG .((((((((((((.(..((..((((((......))))))))..).))))))))))((((......)))).........))........................................ (-23.38 = -22.50 + -0.88)


| Location | 304,395 – 304,510 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.51 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 304395 115 - 27905053 CAAAAGCGCGGGCCCUU-UACACUUGCGUUCGUUCUCCUUCGCGAGUGUGUAUUUGUGGCCGUGCACAA----GUGCAUUUGUGUGUGUGCACUCGGGAUUCCUCUUCCAUUCCAAUCGC .......((((((((..-((((((((((............)))))))))).....).))))(((((((.----(..(....)..).)))))))..((((......))))........))) ( -40.20) >DroSec_CAF1 39607 115 - 1 CAAAAGCGCGGGCCCUU-UACACUUGCGUUCGUUCUCCUUCGCGAGUGCGCAUUUGUGACCGUGCACGA----GUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCUAAUUGC .....((((((......-..((((((((............))))))))(((....))).))))))..((----((((((........))))))))((((......))))........... ( -40.50) >DroEre_CAF1 44025 120 - 1 CAAAAGCGCGGGCCCUUCUACACUUGCGUUCGUUCUCUUUCGCGGGUGCGUAUUUGUGGCCGUGAACGAGUGAGUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCCAAUUGC ....((((((((..........))))))))....(((.(((((((.((((....)))).))))))).))).((((((((........))))))))((((......))))........... ( -40.60) >DroYak_CAF1 43978 116 - 1 UAAAAGCGCGGGCCCUUCUACACUUGCGUUCGUUCUCCUUCGCGAGUGCGUAUUUGUGGCCGUGUACGA----GUGCAUUCGUAUGUGUGCACACGGAAUUCCUCUUCCAUUCCAAUUGC .......((((((((.....((((((((............)))))))).......).))))(((((((.----((((....)))).)))))))..((((......))))........))) ( -34.50) >consensus CAAAAGCGCGGGCCCUU_UACACUUGCGUUCGUUCUCCUUCGCGAGUGCGUAUUUGUGGCCGUGCACGA____GUGCAUUCGUGUGUGUGCACUCGGAAUUCCUCUUCCAUUCCAAUUGC .....((((((.(.(.....((((((((............)))))))).......).).))))))........((((((........))))))..((((......))))........... (-31.75 = -31.50 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:14 2006