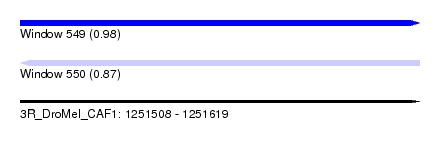
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,251,508 – 1,251,619 |
| Length | 111 |
| Max. P | 0.982031 |

| Location | 1,251,508 – 1,251,619 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -49.15 |
| Consensus MFE | -34.97 |
| Energy contribution | -36.60 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1251508 111 + 27905053 AGCUCCUGCUCCUGCUCCCCCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCCUAUCCCGAUCCUGGAGGCGCUUCUUCUGGAGGUGGUAGCUGCAGGAGCAGGCC .((((((((..((((..((((((..((.....))..))))(((..(((((((((.(((............))))))))))))..))).))..))))..))))))))..... ( -55.80) >DroSec_CAF1 26759 108 + 1 AGCUCCUGCUCCUGCUCCCUCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCGUAUCCCGAUUCUGAAGGCGCU---UCUGGAGGUGGUAGCUGCAGGAGCAGGCC .((((((((..((((..((((((..((.....))..))......((((((((((.(((...........))).))))))---))))))))..))))..))))))))..... ( -55.40) >DroEre_CAF1 26106 110 + 1 -GCUCCUGCUCCUGCUCCUCCGCAAGGAUCCUUCGCUCGGUCAUCAGAGGCGCCAUCUUAUCCCGAUCCUGGAGGCGCAUUUUCUGGAGGUGGUAGCUGCAAGAGCAGGCC -...(((((((.(((.(((((....))).(((((.(((........)))(((((.(((............)))))))).......))))).)).....))).))))))).. ( -42.80) >DroYak_CAF1 31054 104 + 1 -G------CGCAUGCUCCUCCGGAAAGAUCCUUCGCUCGGUCAUCAGAGGCAUCCUCUGAUCUCGAUCCUGGGGACGCUUCGUCUGGAGGUGGUAACUGCAAGAGCAGGCC -.------.((.((((((((((((..((((........))))....(((((.(((((.(((....)))..))))).))))).)))))))).)))).((((....)))))). ( -42.60) >consensus _GCUCCUGCUCCUGCUCCCCCGAAAGGAUCCUCCGCUCGGUCAUCAGAGGCGCCCUCUUAUCCCGAUCCUGGAGGCGCUUCUUCUGGAGGUGGUAGCUGCAAGAGCAGGCC ....(((((((((((.((.((....))..(((((.(((........)))(((((.(((............)))))))).......))))).)).....))))))))))).. (-34.97 = -36.60 + 1.63)

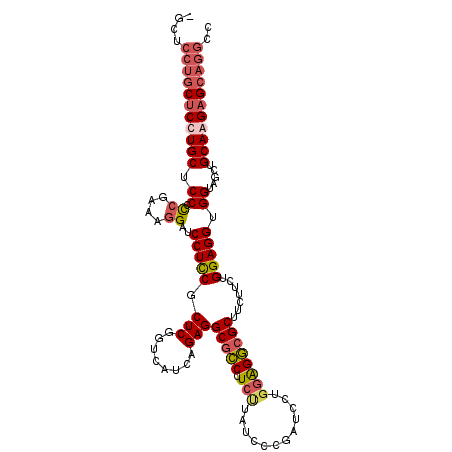

| Location | 1,251,508 – 1,251,619 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -46.64 |
| Consensus MFE | -36.65 |
| Energy contribution | -37.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1251508 111 - 27905053 GGCCUGCUCCUGCAGCUACCACCUCCAGAAGAAGCGCCUCCAGGAUCGGGAUAGGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUUCGGGGGAGCAGGAGCAGGAGCU ..(((((((((((.(....).(((((..(((.....(((((....((((....(((((...))))).....))))..)))))...)))..))))).))))))))))).... ( -54.30) >DroSec_CAF1 26759 108 - 1 GGCCUGCUCCUGCAGCUACCACCUCCAGA---AGCGCCUUCAGAAUCGGGAUACGAGGGCGCCUCUGAUGACCGAGCGGAGGAUCCUUUCGAGGGAGCAGGAGCAGGAGCU ..(((((((((((........(((((...---.((((((((.............))))))))(((........))).))))).((((.....))))))))))))))).... ( -52.72) >DroEre_CAF1 26106 110 - 1 GGCCUGCUCUUGCAGCUACCACCUCCAGAAAAUGCGCCUCCAGGAUCGGGAUAAGAUGGCGCCUCUGAUGACCGAGCGAAGGAUCCUUGCGGAGGAGCAGGAGCAGGAGC- ..(((((((((((.((.................)).(((((....((((....(((.(....)))).....))))((.(((....)))))))))).)))))))))))...- ( -43.53) >DroYak_CAF1 31054 104 - 1 GGCCUGCUCUUGCAGUUACCACCUCCAGACGAAGCGUCCCCAGGAUCGAGAUCAGAGGAUGCCUCUGAUGACCGAGCGAAGGAUCUUUCCGGAGGAGCAUGCG------C- .((.(((((((((........(((...((((...))))...))).(((..((((((((...))))))))...)))))...(((....)))..))))))).)).------.- ( -36.00) >consensus GGCCUGCUCCUGCAGCUACCACCUCCAGAAGAAGCGCCUCCAGGAUCGGGAUAAGAGGGCGCCUCUGAUGACCGAGCGAAGGAUCCUUUCGGAGGAGCAGGAGCAGGAGC_ ..(((((((((((........(((((.......((((((.(.............).))))))(((........))).))))).((((.....))))))))))))))).... (-36.65 = -37.65 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:31 2006