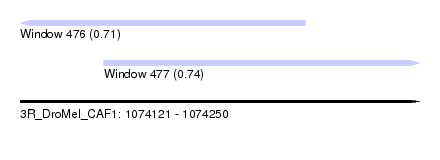
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,074,121 – 1,074,250 |
| Length | 129 |
| Max. P | 0.741237 |

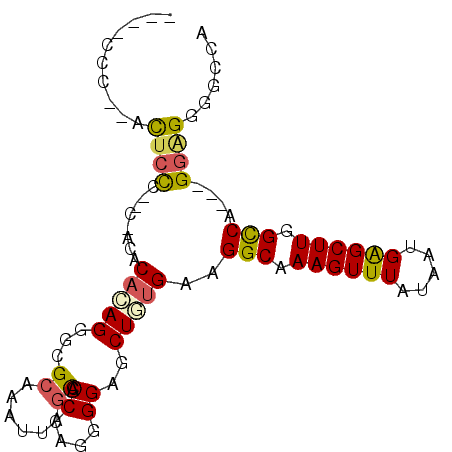
| Location | 1,074,121 – 1,074,213 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.00 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1074121 92 - 27905053 AUACCCC--ACUGCCCCCCACACACAGGGUGCAAAUUAGCACCCAGGGGAGCUGUGAAGGCAAAGUUUAUAAUGAGCUUGGCCA---GGAGGGGCCA .......--...(((((((...(((.((((((......)))))).((....)))))..(((.((((((.....)))))).))).---)).))))).. ( -39.20) >DroSec_CAF1 10196 87 - 1 AUACCCC--ACUCCC-----CACACAGGGCGCAAAUUAGCACCCAGGGGAGCUGUGAAGGCAAAGUUUAUAAUGAGCUUGGCCA---GGAGGGGCCA ...((((--.(((((-----(.....(((.((......)).))).)))))).......(((.((((((.....)))))).))).---...))))... ( -35.70) >DroSim_CAF1 10312 87 - 1 AUACCCC--ACUCCC-----CACACAGGGCGCAAAUUAGCACCCAGGGGAGCUGUGAAGGCAAAGUUUAUAAUGAGCUUGGCCA---GGAGGGGCCA ...((((--.(((((-----(.....(((.((......)).))).)))))).......(((.((((((.....)))))).))).---...))))... ( -35.70) >DroEre_CAF1 10530 85 - 1 -----CC--ACUCCC--GGACACAGAGAACGCAAAUUAGCACCCAGGGGAGCUGUGAAGGCAAAGUUUAUAAUGGGCUUGGCCA---GGGGGGUCCA -----..--((((((--...(((((.....((......)).((....))..)))))..(((.((((((.....)))))).))).---.))))))... ( -28.80) >DroYak_CAF1 16389 85 - 1 -----CG--CCGCCC--CUAUACAGAGGGCGCAAAUUACUACCCAGUGGAACUGUGAAGGCAAAGUUUAUAAUGAGCUUGGCCA---GGGGGGUCCA -----.(--((.(((--((..........((((..(((((....)))))...))))..(((.((((((.....)))))).))))---)))))))... ( -27.00) >DroAna_CAF1 11257 91 - 1 ----CCCGGAUUCUU--CGGCACAGAGGGCGCAAAUUAGCACCCAGGGGAGCUUCGUGGGCAAAGUUUAUAAUGAGCUUGGUCAUAAGGGGGGUCCA ----...((((((((--(..(((...(((.((......)).)))(((....))).)))(((.((((((.....)))))).)))....))))))))). ( -30.90) >consensus ____CCC__ACUCCC__C_ACACACAGGGCGCAAAUUAGCACCCAGGGGAGCUGUGAAGGCAAAGUUUAUAAUGAGCUUGGCCA___GGAGGGGCCA ..........((((........(((((...((......)).((....))..)))))..(((.((((((.....)))))).)))....))))...... (-19.02 = -19.00 + -0.02)

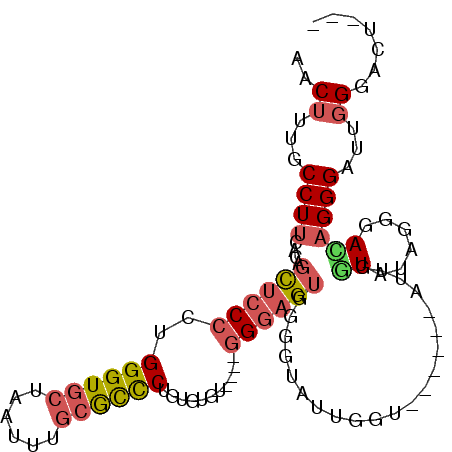
| Location | 1,074,148 – 1,074,250 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.05 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1074148 102 + 27905053 AACUUUGCCUUCACAGCUCCCCUGGGUGCUAAUUUGCACCCUGUGUGUGGGGGGCAGUGGGGUAUUGGUAUUGGUAUUGGUAUAGGAAAAGGGACUUGGAUU--- ..((((.(((((((.(((((((.((((((......)))))).......))))))).)))).((((..(((....)))..))))))).))))...........--- ( -39.60) >DroSec_CAF1 10223 91 + 1 AACUUUGCCUUCACAGCUCCCCUGGGUGCUAAUUUGCGCCCUGUGUG-----GGGAGUGGGGUAUUGGU------AUUGGUAUAGGGACAGGGAUUGGGACU--- ..(((((((((..(.(((((((.((((((......)))))).....)-----)))))).).(((((...------...))))))))).))))).........--- ( -33.70) >DroSim_CAF1 10339 91 + 1 AACUUUGCCUUCACAGCUCCCCUGGGUGCUAAUUUGCGCCCUGUGUG-----GGGAGUGGGGUAUUGGU------AUUGGUAUAGGGACAGGGAUUGGGACU--- ..(((((((((..(.(((((((.((((((......)))))).....)-----)))))).).(((((...------...))))))))).))))).........--- ( -33.70) >DroEre_CAF1 10557 78 + 1 AACUUUGCCUUCACAGCUCCCCUGGGUGCUAAUUUGCGUUCUCUGUGUCC--GGGAGUGG------------------GGUAUUGGGAUUGGGAUUGG------- ..(..(.((.((.(((..((((.(..(((......)))..)((((....)--)))...))------------------))..))).))..)).)..).------- ( -20.80) >DroYak_CAF1 16416 87 + 1 AACUUUGCCUUCACAGUUCCACUGGGUAGUAAUUUGCGCCCUCUGUAUAG--GGGCGGCG----------------UUGCUAUAAUGAGAGGGAUUGGGGCUCGU ......((((...(((((((.((..((((((((...((((((.......)--)))))..)----------------)))))))....)).))))))))))).... ( -31.30) >consensus AACUUUGCCUUCACAGCUCCCCUGGGUGCUAAUUUGCGCCCUGUGUGU____GGGAGUGGGGUAUUGGU______AUUGGUAUAGGGACAGGGAUUGGGACU___ ..((...((((....((((((..((((((......))))))...........)))))).....................((......))))))...))....... (-13.14 = -13.98 + 0.84)

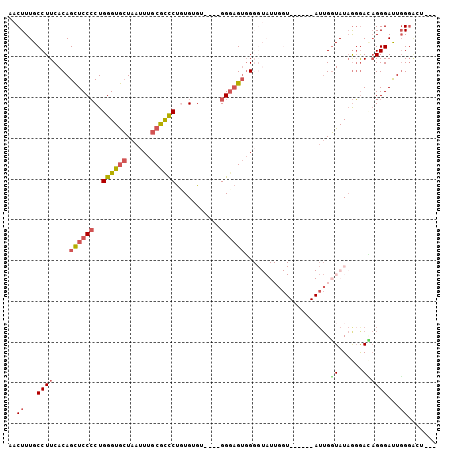
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:18 2006