
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 849,896 – 850,200 |
| Length | 304 |
| Max. P | 0.988845 |

| Location | 849,896 – 850,016 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -37.04 |
| Energy contribution | -37.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849896 120 + 27905053 AGACCCGACCAGGCUGUCUGCCGCUUUGUCAGACAACCCUUGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGA ..(((((((.((((.(....).)))).)))......((((.(((((......)))))(((((...((((.....))))(((((((((.......))))))))).))))).)))))))).. ( -42.50) >DroSec_CAF1 25545 119 + 1 AGACCCGACCAGCCUGUCUGCCGCUCUGUCAGACAACCCUUGGAGAUUGUUUUCUUCUGCAGGCCCGA-AACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGA ..((((........((((((.(.....).)))))).((((.(((((......)))))(((((...(((-(....))))(((((((((.......))))))))).))))).)))))))).. ( -41.10) >DroSim_CAF1 24776 120 + 1 AGACCCGACCAGCCUGUCUGCCGCUCUGUCAGACAACCCUUGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGA ..((((........((((((.(.....).)))))).((((.(((((......)))))(((((...((((.....))))(((((((((.......))))))))).))))).)))))))).. ( -41.10) >DroEre_CAF1 26507 119 + 1 AUACCCGACCAGCCUGUCUGCCGGUCUGUCAGACAACCCUUGAAGAUUGUUUUCUUCUGCCAGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGG-GGGUGG .(((((((((.((......)).))))((((((.........(((((......))))).....................(((((((((.......))))))))).))).)))..-))))). ( -36.70) >consensus AGACCCGACCAGCCUGUCUGCCGCUCUGUCAGACAACCCUUGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGA ..((((........((((((.(.....).)))))).((((.(((((......)))))(((((...((((.....))))(((((((((.......))))))))).))))).)))))))).. (-37.04 = -37.85 + 0.81)

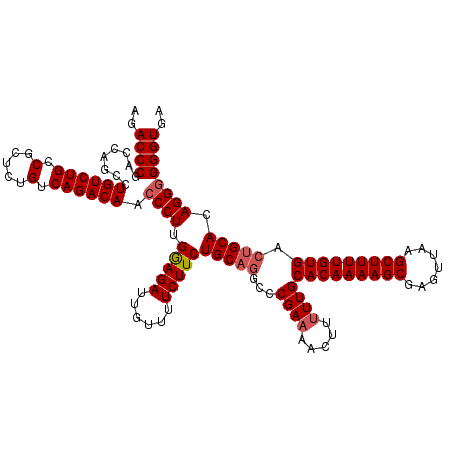
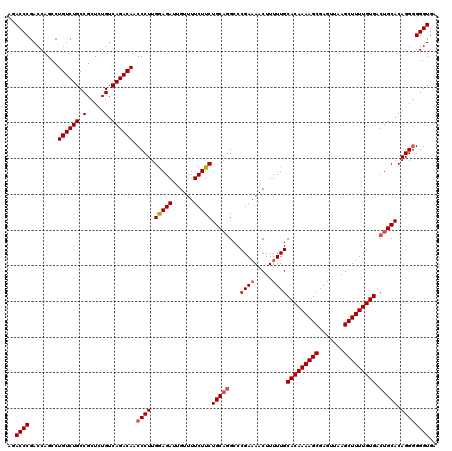
| Location | 849,936 – 850,046 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -28.41 |
| Energy contribution | -33.60 |
| Covariance contribution | 5.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849936 110 + 27905053 UGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGACUAUCCGUGGUAAAGGGGUGGAGCAGGGGA---------- ............((((((((..((((............(((((((((.......)))))))))..(((.((.((((....))..)).)))))...))))...))))))))---------- ( -37.60) >DroSec_CAF1 25585 119 + 1 UGGAGAUUGUUUUCUUCUGCAGGCCCGA-AACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGACUAUCUGGGGUAGAGGGGUUGAGCAGGGGAGGAGGGGCAG ....(.((.(((((((((((.(((((..-.........(((((((((.......))))))))).((((.(((..((....))..)))..))))..)))))..))))))))))).)).).. ( -48.10) >DroSim_CAF1 24816 118 + 1 UGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGACUAUCUGGGGUAGAGGGGUUGAGCAGGGGAGGAGGGGC-- ......((.(((((((((((.(((((............(((((((((.......))))))))).((((.(((..((....))..)))..))))..)))))..))))))))))).))..-- ( -47.90) >DroEre_CAF1 26547 105 + 1 UGAAGAUUGUUUUCUUCUGCCAGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGG-GGGUGGCUAUCCGCGGUGAGGGGGUA----------AGAGGG---- .(((((......))))).((((.(((............(((((((((.......))))))))).((....)))-)).))))(((((.(.....).)))))----------......---- ( -37.30) >consensus UGGAGAUUGUUUUCUUCUGCAGGCCCGAAAACUUUUUGCACAAAAGCGAGUUAAGCUUUUGUGACUGCACAGGGGGGUGACUAUCCGGGGUAAAGGGGUUGAGCAGGGGAGGAGGG____ .........(((((((((((..((((............(((((((((.......))))))))).((((...(((((....)).)))...))))..))))...)))))))))))....... (-28.41 = -33.60 + 5.19)

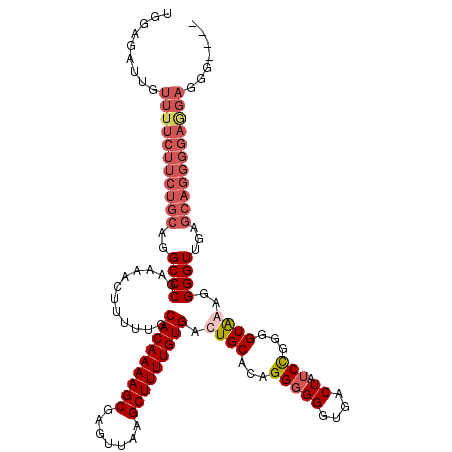
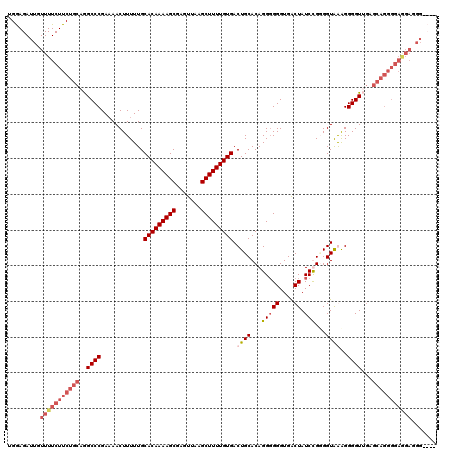
| Location | 849,936 – 850,046 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849936 110 - 27905053 ----------UCCCCUGCUCCACCCCUUUACCACGGAUAGUCACCCCCCUGUGCAGUCACAAAAGCUUAACUCGCUUUUGUGCAAAAAGUUUUCGGGCCUGCAGAAGAAAACAAUCUCCA ----------((..((((....(((......(((((............)))))..(.(((((((((.......))))))))))...........)))...))))..))............ ( -25.30) >DroSec_CAF1 25585 119 - 1 CUGCCCCUCCUCCCCUGCUCAACCCCUCUACCCCAGAUAGUCACCCCCCUGUGCAGUCACAAAAGCUUAACUCGCUUUUGUGCAAAAAGUU-UCGGGCCUGCAGAAGAAAACAAUCUCCA (((((((.......((((.((........((........))........)).)))).(((((((((.......))))))))).........-..)))...))))................ ( -23.69) >DroSim_CAF1 24816 118 - 1 --GCCCCUCCUCCCCUGCUCAACCCCUCUACCCCAGAUAGUCACCCCCCUGUGCAGUCACAAAAGCUUAACUCGCUUUUGUGCAAAAAGUUUUCGGGCCUGCAGAAGAAAACAAUCUCCA --........((..((((....(((..............(.(((......)))).(.(((((((((.......))))))))))...........)))...))))..))............ ( -23.30) >DroEre_CAF1 26547 105 - 1 ----CCCUCU----------UACCCCCUCACCGCGGAUAGCCACCC-CCUGUGCAGUCACAAAAGCUUAACUCGCUUUUGUGCAAAAAGUUUUCGGGCUGGCAGAAGAAAACAAUCUUCA ----......----------.....((.......))...((((.((-(....((...(((((((((.......)))))))))......))....))).)))).(((((......))))). ( -27.40) >consensus ____CCCUCCUCCCCUGCUCAACCCCUCUACCCCAGAUAGUCACCCCCCUGUGCAGUCACAAAAGCUUAACUCGCUUUUGUGCAAAAAGUUUUCGGGCCUGCAGAAGAAAACAAUCUCCA ..............((((....(((......(((((............)))))..(.(((((((((.......))))))))))...........)))...))))................ (-19.14 = -20.32 + 1.19)

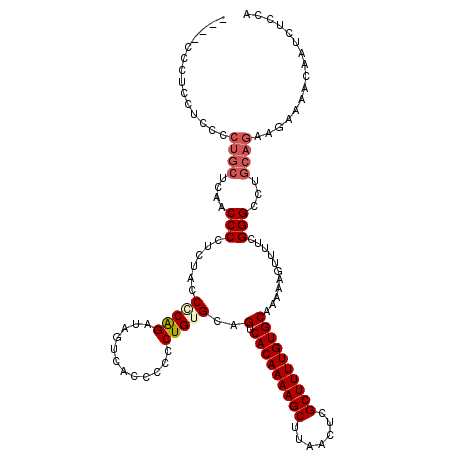
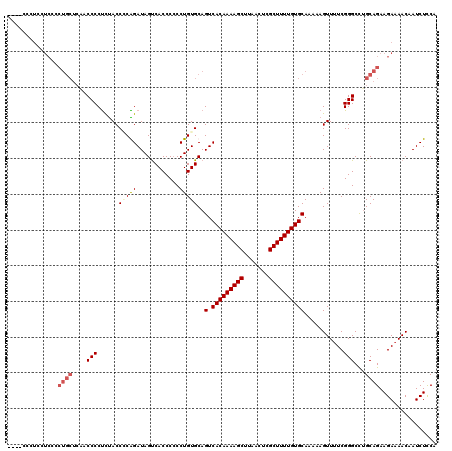
| Location | 850,016 – 850,120 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -22.25 |
| Energy contribution | -24.38 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 850016 104 + 27905053 CUAUCCGUGGUAAAGGGGUGGAGCAGGGGA----------------AGAGGAUACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCA ((((((.(.....).)))))).((((((..----------------...((......(((....))).....)).....))).)))..(((....)))(((((((((...))))))))). ( -33.10) >DroSec_CAF1 25664 120 + 1 CUAUCUGGGGUAGAGGGGUUGAGCAGGGGAGGAGGGGCAGAGGGGCAGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCA ....(((..((((((..((((.((............((......))...(....)..(((....)))....))))))..))).))).)))........(((((((((...))))))))). ( -34.90) >DroSim_CAF1 24896 112 + 1 CUAUCUGGGGUAGAGGGGUUGAGCAGGGGAGGAGGGGC--------AGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCA ....(((..((((((..((((.((...(..((..(.((--------...(....)...))..)..))..).))))))..))).))).)))........(((((((((...))))))))). ( -36.20) >DroEre_CAF1 26626 100 + 1 CUAUCCGCGGUGAGGGGGUA----------AGAGGG----------ACAAGGGACAAGGCGACAGCUGACAGCCAACAGCUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCA ..........((((((.(((----------((.(((----------.....((....(((....))).....)).....)))((((.((((....))))...))))..))))).)))))) ( -29.80) >consensus CUAUCCGGGGUAAAGGGGUUGAGCAGGGGAGGAGGG__________AGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCA .........(((...((((((............................(....)..(((...........)))..)))))).)))..(((....)))(((((((((...))))))))). (-22.25 = -24.38 + 2.13)

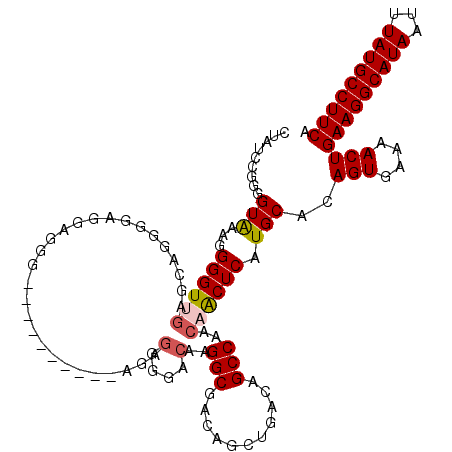

| Location | 850,046 – 850,160 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -30.10 |
| Energy contribution | -32.35 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 850046 114 + 27905053 ------AGAGGAUACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCC ------...(((.....(((...........)))........((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).)))))....)))... ( -29.30) >DroSec_CAF1 25704 120 + 1 AGGGGCAGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCC .((((..(((.((....(((....))).....)).....)))((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).))))).....)))). ( -38.10) >DroSim_CAF1 24934 114 + 1 ------AGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCC ------...(((((...(((...........)))........((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).)))))....))))). ( -35.80) >DroEre_CAF1 26652 114 + 1 ------ACAAGGGACAAGGCGACAGCUGACAGCCAACAGCUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCU ------....((((.(((.....(((((........))))).((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).)))))))).)))).. ( -33.60) >consensus ______AGAGGGGACAAGGCGACAGCUGACAGCCAACAACUCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCC .........(((((...(((...........)))........((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).)))))....))))). (-30.10 = -32.35 + 2.25)


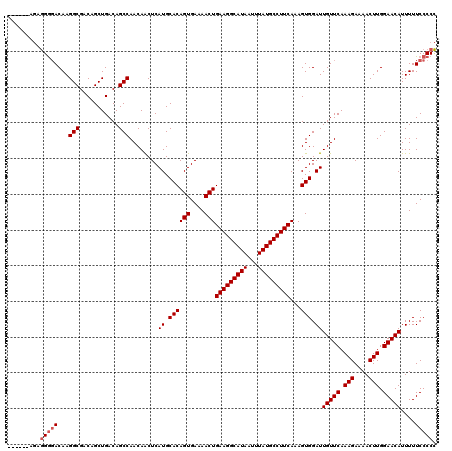
| Location | 850,046 – 850,160 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -33.75 |
| Energy contribution | -33.25 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 850046 114 - 27905053 GGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGAGUUGUUGGCUGUCAGCUGUCGCCUUGUAUCCUCU------ (((((((((.(((((.(((....))).))))).......(((((((((((...))))))))))))))))...(((((.(.((...((((.....))))..))).))))))))).------ ( -33.00) >DroSec_CAF1 25704 120 - 1 GGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGAGUUGUUGGCUGUCAGCUGUCGCCUUGUCCCCUCUGCCCCU (((((((((.(((((.(((....))).))))).......(((((((((((...)))))))))))))))).....(((.(((.....(((.(.(....).)))).......)))))))))) ( -35.60) >DroSim_CAF1 24934 114 - 1 GGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGAGUUGUUGGCUGUCAGCUGUCGCCUUGUCCCCUCU------ (((((((((.(((((.(((....))).))))).......(((((((((((...))))))))))))))))...(((.((.(((((........)))))))))).....))))...------ ( -35.10) >DroEre_CAF1 26652 114 - 1 AGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGAGCUGUUGGCUGUCAGCUGUCGCCUUGUCCCUUGU------ ((((((....(((((.(((....))).)))))..........(((((.....((((((...((((....)))).))))))((((........))))....))))).))))))..------ ( -34.80) >consensus GGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGAGUUGUUGGCUGUCAGCUGUCGCCUUGUCCCCUCU______ (((((((((.(((((.(((....))).))))).......(((((((((((...))))))))))))))))...(((.((.(((((........)))))))))).....))))......... (-33.75 = -33.25 + -0.50)

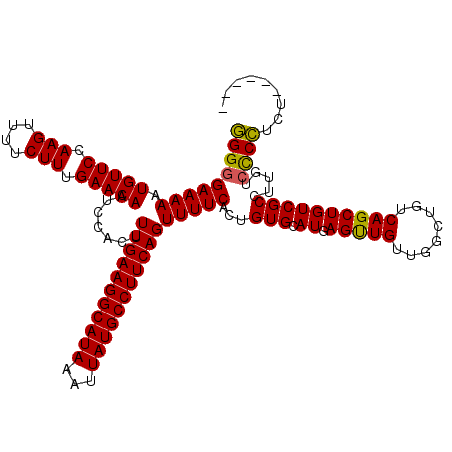
| Location | 850,080 – 850,200 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.37 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 850080 120 + 27905053 UCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCAGAGCAGCCAGAGCCGC ...(((...((((....((((((((((...))))))))))..((...((((((.(((....)))((((.....))))...))))))..)).))))..)))......((.((....)).)) ( -29.80) >DroSec_CAF1 25744 111 + 1 UCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCA---------GAGCCGC ...(((...((((....((((((((((...))))))))))..((...((((((.(((....)))((((.....))))...))))))..)).))))..)))....---------....... ( -27.80) >DroSim_CAF1 24968 111 + 1 UCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCA---------GAGCCGC ...(((...((((....((((((((((...))))))))))..((...((((((.(((....)))((((.....))))...))))))..)).))))..)))....---------....... ( -27.80) >DroEre_CAF1 26686 111 + 1 UCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCUGAACAACAACAUCACCACCAGCCA---------GAGGAGC ..((.((((((....)))(((((((((...)))))))))...))).))(((((.(((....))).)))))...((((.(((.....................))---------).)))). ( -27.70) >consensus UCAUGCACAGUGAAAACUGAAGGCAUAAUUUAUGCCUUCAAAGUGGAUUGUUCAAAGAAAACUUGGAACAUUUUUCCCCCGAACAACAACAUCACCAGCAGCCA_________GAGCCGC ...(((...((((....((((((((((...))))))))))..((...((((((.(((....)))((((.....))))...))))))..)).))))..))).................... (-26.40 = -26.65 + 0.25)

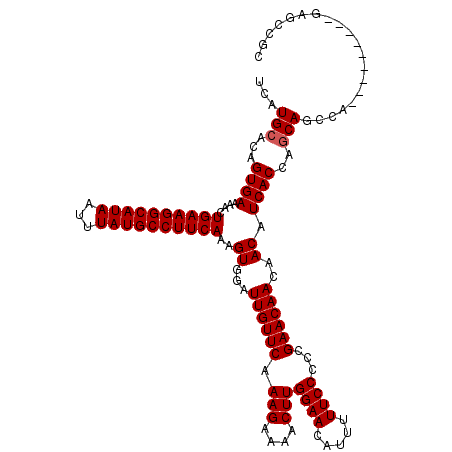
| Location | 850,080 – 850,200 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.37 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -35.64 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 850080 120 - 27905053 GCGGCUCUGGCUGCUCUGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGA (((((....)))))......(((((((((....(((((((((((((((..((...))..))))))))))))))).....(((((((((((...)))))))))))...)))))).)))... ( -43.00) >DroSec_CAF1 25744 111 - 1 GCGGCUC---------UGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGA (((((..---------..))))).(((((....(((((((((((((((..((...))..))))))))))))))).....(((((((((((...)))))))))))...)))))........ ( -39.70) >DroSim_CAF1 24968 111 - 1 GCGGCUC---------UGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGA (((((..---------..))))).(((((....(((((((((((((((..((...))..))))))))))))))).....(((((((((((...)))))))))))...)))))........ ( -39.70) >DroEre_CAF1 26686 111 - 1 GCUCCUC---------UGGCUGGUGGUGAUGUUGUUGUUCAGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGA .......---------..((((((((.(((...(((((((((((((((..((...))..)))))))))))))))......((((((((((...))))))))))))).)))))).)).... ( -35.40) >consensus GCGGCUC_________UGGCUGCUGGUGAUGUUGUUGUUCGGGGGAAAAAUGUUCCAAGUUUUCUUUGAACAAUCCACUUUGAAGGCAUAAAUUAUGCCUUCAGUUUUCACUGUGCAUGA ((((((...........)))))).(((((....(((((((((((((((..((...))..))))))))))))))).....(((((((((((...)))))))))))...)))))........ (-35.64 = -36.20 + 0.56)

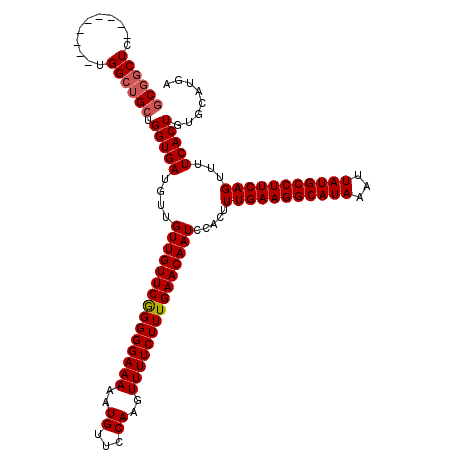
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:54 2006