
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,432,344 – 5,432,461 |
| Length | 117 |
| Max. P | 0.999908 |

| Location | 5,432,344 – 5,432,461 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.53 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -37.90 |
| Energy contribution | -37.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
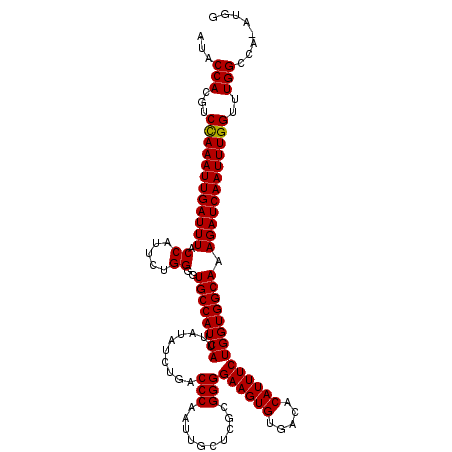
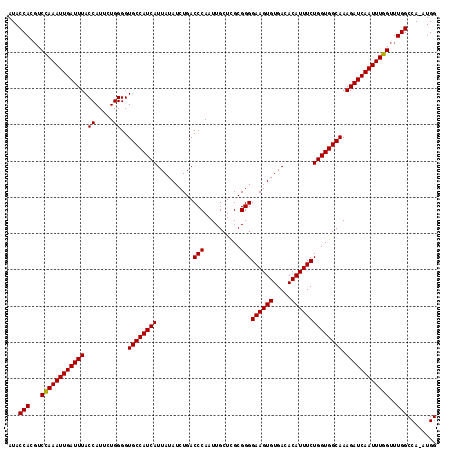
>3R_DroMel_CAF1 5432344 117 + 27905053 AUACCAAGUCCAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUCGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCA-AUGG ...(((((.((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).)))))))))))))))))...-.... ( -40.70) >DroSec_CAF1 18976 117 + 1 AUACCACGUCCAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUCGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCA-AUGG ...(((...((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).))))))))))))..)))...-.... ( -38.20) >DroSim_CAF1 14000 117 + 1 AUACCACGUCCAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUCGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCA-AUGG ...(((...((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).))))))))))))..)))...-.... ( -38.20) >DroEre_CAF1 19570 117 + 1 AUACCAGCUCCAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUUGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCA-CUGG ...((((..((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).)))))))))))).))))...-.... ( -39.00) >DroYak_CAF1 19895 118 + 1 AUACCACGUCUAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUUGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCAAAUGG ...(((...((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).))))))))))))..)))........ ( -35.90) >consensus AUACCACGUCCAAAUUGAUUUACCAUUCUGGGGUGCCAUCAUUAUAUCUGACCCAAUUGCUCGCGGGGAAGUGUGACACAUUUCUGGUGGCAAAGAUCAAUUUGGUUUGGCCA_AUGG ...(((...((((((((((((.((.....))..((((((((..........(((..........)))((((((.....)))))))))))))).))))))))))))..)))........ (-37.90 = -37.74 + -0.16)

| Location | 5,432,344 – 5,432,461 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.53 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
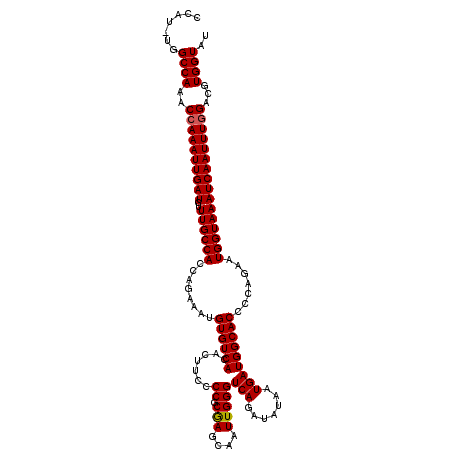
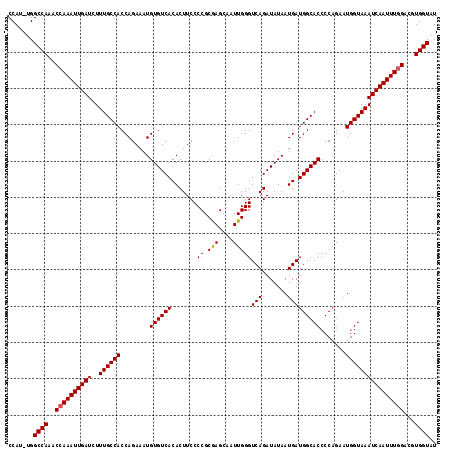
>3R_DroMel_CAF1 5432344 117 - 27905053 CCAU-UGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCGAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUGGACUUGGUAU ....-..(((((.((((((((((..((((((........((((((((((..((((........))))..))......)).)))))).......))))))))))))))))..))))).. ( -32.86) >DroSec_CAF1 18976 117 - 1 CCAU-UGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCGAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUGGACGUGGUAU ....-..((((..((((((((((..((((((........((((((((((..((((........))))..))......)).)))))).......))))))))))))))))...)))).. ( -33.06) >DroSim_CAF1 14000 117 - 1 CCAU-UGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCGAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUGGACGUGGUAU ....-..((((..((((((((((..((((((........((((((((((..((((........))))..))......)).)))))).......))))))))))))))))...)))).. ( -33.06) >DroEre_CAF1 19570 117 - 1 CCAG-UGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCAAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUGGAGCUGGUAU ((((-(.......((((((((((..((((((........((((((((((..((((........))))..))......)).)))))).......)))))))))))))))).)))))... ( -32.16) >DroYak_CAF1 19895 118 - 1 CCAUUUGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCAAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUAGACGUGGUAU .......((((..(.((((((((..((((((........((((((((((..((((........))))..))......)).)))))).......)))))))))))))).)...)))).. ( -26.56) >consensus CCAU_UGGCCAAACCAAAUUGAUCUUUGCCACCAGAAAUGUGUCACACUUCCCCGCGAGCAAUUGGGUCAGAUAUAAUGAUGGCACCCCAGAAUGGUAAAUCAAUUUGGACGUGGUAU .......((((..((((((((((..((((((........((((((.......((.(((....)))))(((.......))))))))).......))))))))))))))))...)))).. (-31.40 = -31.36 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:31 2006