| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 4,799,953 – 4,800,067 |
| Length | 114 |
| Max. P | 0.848300 |

| Location | 4,799,953 – 4,800,067 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -23.35 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
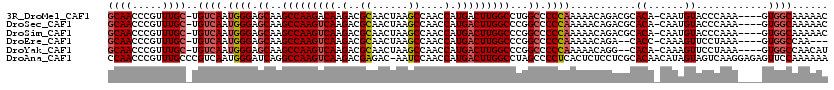
>3R_DroMel_CAF1 4799953 114 + 27905053 GCAACCCGUUUGC-UGUCAAUGGGAGCAAGCCAAGACAAGACGCAACUAAGCCAACCAUGACUUGGCCUGGCCCCCAAAAACAGACGCACA-CAAUGUACCCAAA----GUGGCAAAAAC ((.....((...(-(((...((((.((..((((((.((.(..((......))....).)).))))))...)).))))...))))..)).((-(..((....))..----)))))...... ( -28.50) >DroSec_CAF1 35368 114 + 1 GCAACCCGUUUGC-UGUCAAUGGGAGCAAGCCAAGUCAAGACGCAACUAAGCCAACCAUGACUUGGCCCGGCCCCCAAAAACAGACGCACA-CAAUGUACCCAAA----GUGGCAAAAAC ((.....((...(-(((...((((.((..(((((((((.(..((......))....).)))))))))...)).))))...))))..)).((-(..((....))..----)))))...... ( -33.90) >DroSim_CAF1 31472 114 + 1 GCAACCCGUUUGC-UGUCAAUGGGAGCAAGCCAAGUCAAGACGCAACUAAGCCAACCAUGACUUGGCCCGGCCCCCAAAAACAGACGCACA-CAAUGUACCCAAA----GUGGCAAAAAC ((.....((...(-(((...((((.((..(((((((((.(..((......))....).)))))))))...)).))))...))))..)).((-(..((....))..----)))))...... ( -33.90) >DroEre_CAF1 36206 109 + 1 GCAACCCGUUUGC-UGUCAAUGGGAGCAAGCCAAGUCAAGACGCAACUAAGCCAACCAUGACUUGGCCCGGCCCCCAAAAACAGA--CACC-CAAAGUUCCUAAA----GUGGCCAA--- .......(..(((-(((...((((.((..(((((((((.(..((......))....).)))))))))...)).))))...)))).--))..-)...((..(....----)..))...--- ( -31.10) >DroYak_CAF1 36491 112 + 1 GCAACCCGUUUGC-UGUCAAUGGGAGCAAGCCAAGUCAAGACGCAACUAAGCCAACCAUGACUUGGCCCGGCCCCCAAAAACAGG--CACA-CAAAGUUCCUAAA----GUGGCCAACAU ((((.....))))-(((...((((.((..(((((((((.(..((......))....).)))))))))...)).))))...)))((--(.((-(............----))))))..... ( -35.20) >DroAna_CAF1 35134 119 + 1 CCAACCCGUUUGCCCGUCAAUGGGAUCAGGCCAAGUCAAGACGAGAC-AAUCCAACCAUGACUUGGCCUAGCCCCUCACUCUCCUCGCACAACAUAGUAGUCAAGGAGAGUUCCAAAAAA ....((((((........))))))...(((((((((((.(..((...-..))....).)))))))))))........((((((((.((((......)).))..))))))))......... ( -36.50) >consensus GCAACCCGUUUGC_UGUCAAUGGGAGCAAGCCAAGUCAAGACGCAACUAAGCCAACCAUGACUUGGCCCGGCCCCCAAAAACAGACGCACA_CAAAGUACCCAAA____GUGGCAAAAAC ((((.....))))..((((.((((.((..(((((((((.(..((......))....).)))))))))...)).))))...........((......))............))))...... (-23.35 = -23.93 + 0.58)
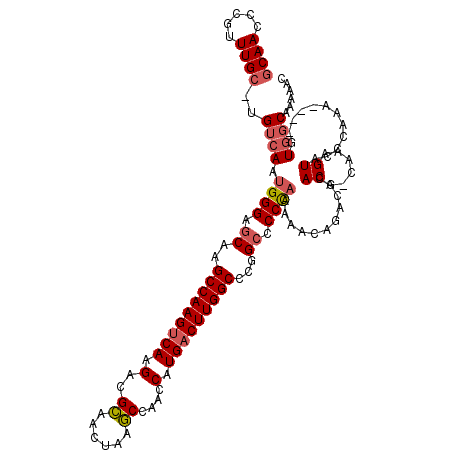
| Location | 4,799,953 – 4,800,067 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -47.48 |
| Consensus MFE | -33.77 |
| Energy contribution | -34.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 4799953 114 - 27905053 GUUUUUGCCAC----UUUGGGUACAUUG-UGUGCGUCUGUUUUUGGGGGCCAGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGUCUUGGCUUGCUCCCAUUGACA-GCAAACGGGUUGC ((((..(((((----..((....))..)-)).))(.(((((..(((((((.((((((((.((.((...((......))..)))).)))))))))))))))..))))-))))))....... ( -45.80) >DroSec_CAF1 35368 114 - 1 GUUUUUGCCAC----UUUGGGUACAUUG-UGUGCGUCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA-GCAAACGGGUUGC ((((..(((((----..((....))..)-)).))(.(((((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))))-))))))....... ( -48.90) >DroSim_CAF1 31472 114 - 1 GUUUUUGCCAC----UUUGGGUACAUUG-UGUGCGUCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA-GCAAACGGGUUGC ((((..(((((----..((....))..)-)).))(.(((((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))))-))))))....... ( -48.90) >DroEre_CAF1 36206 109 - 1 ---UUGGCCAC----UUUAGGAACUUUG-GGUG--UCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA-GCAAACGGGUUGC ---....((..----....))(((((.(-..((--.(((((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))))-)))..)))))).. ( -46.00) >DroYak_CAF1 36491 112 - 1 AUGUUGGCCAC----UUUAGGAACUUUG-UGUG--CCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA-GCAAACGGGUUGC .......((..----....))(((((.(-(.((--(.((((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..))))-))).))))))).. ( -45.40) >DroAna_CAF1 35134 119 - 1 UUUUUUGGAACUCUCCUUGACUACUAUGUUGUGCGAGGAGAGUGAGGGGCUAGGCCAAGUCAUGGUUGGAUU-GUCUCGUCUUGACUUGGCCUGAUCCCAUUGACGGGCAAACGGGUUGG .........((((((((((.(.((......)))))))))))))..(((..((((((((((((.((..((...-..))..)).))))))))))))..))).....((......))...... ( -49.90) >consensus GUUUUUGCCAC____UUUGGGUACAUUG_UGUGCGUCUGUUUUUGGGGGCCGGGCCAAGUCAUGGUUGGCUUAGUUGCGUCUUGACUUGGCUUGCUCCCAUUGACA_GCAAACGGGUUGC ..............................((.....((((..(((((((.(((((((((((.((...((......))..))))))))))))))))))))..)))).....))....... (-33.77 = -34.47 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:30 2006