| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 803,214 – 803,334 |
| Length | 120 |
| Max. P | 0.938065 |

| Location | 803,214 – 803,334 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 803214 120 + 27905053 AUGCUAAUCAGCUUUGAUGCCCCGAAGCGCCGGAGCUCCAAGACCUGGAUGUCAGGCCUUCUGGAUUUUGGCCACGAGGGUUGCUUGGCCAUUUCUUAGUAAACUGGGGCAUGGCAAAUU .((((((((......)))(((((((((.(((.((..((((.....))))..)).))))))).(((...((((((.(.......).))))))..))).........))))).))))).... ( -43.60) >DroEre_CAF1 24705 112 + 1 AUGCUAAUCAGCUUCGAUGCCCU--------AGAGCUCCAAGCCUUGAAUGUCAGUCCUUCUGGAUUUUGGCCACGAGGGUUGCUCGGCCAUUUCGUGGUAAACUGGGGCAUGGCAAAUU .((((((((......)))(((((--------((.....(((.(((((...((((((((....))))..))))..))))).)))....(((((...)))))...))))))).))))).... ( -39.20) >DroYak_CAF1 30347 93 + 1 AUGCUAAUCAGCUUCGAUGCCCU--------GG---------------AAGUCAGUCCUUCUGGAUUUUGGCCAAGAGGGUUGCUGGGCCA----GUGGUAAACUGGGGCAUGAAAAAUU .......(((((((((.((((((--------((---------------.((.((..(((..(((.......)))..)))..))))...)))----).))))...)))))).)))...... ( -33.10) >consensus AUGCUAAUCAGCUUCGAUGCCCU________GGAGCUCCAAG_C_UG_AUGUCAGUCCUUCUGGAUUUUGGCCACGAGGGUUGCUCGGCCAUUUCGUGGUAAACUGGGGCAUGGCAAAUU ..(((....)))....(((((((...........................(.(((.((((((((.......))).)))))))))...((((.....)))).....)))))))........ (-24.79 = -24.90 + 0.11)

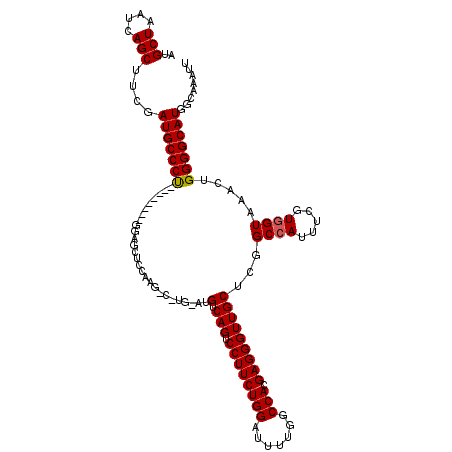
| Location | 803,214 – 803,334 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -20.16 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 803214 120 - 27905053 AAUUUGCCAUGCCCCAGUUUACUAAGAAAUGGCCAAGCAACCCUCGUGGCCAAAAUCCAGAAGGCCUGACAUCCAGGUCUUGGAGCUCCGGCGCUUCGGGGCAUCAAAGCUGAUUAGCAU ..((((..(((((((..............((((((...........))))))........((((((((.....))))))))(((((......))))))))))))))))(((....))).. ( -42.20) >DroEre_CAF1 24705 112 - 1 AAUUUGCCAUGCCCCAGUUUACCACGAAAUGGCCGAGCAACCCUCGUGGCCAAAAUCCAGAAGGACUGACAUUCAAGGCUUGGAGCUCU--------AGGGCAUCGAAGCUGAUUAGCAU ........((((..((((((.....((..((((((((.....)))..)))))...)).....))))))........(((((.(((((..--------..))).)).))))).....)))) ( -29.10) >DroYak_CAF1 30347 93 - 1 AAUUUUUCAUGCCCCAGUUUACCAC----UGGCCCAGCAACCCUCUUGGCCAAAAUCCAGAAGGACUGACUU---------------CC--------AGGGCAUCGAAGCUGAUUAGCAU .....(((((((((...........----(((((.((.......)).))))).......((((......)))---------------).--------.)))))).)))(((....))).. ( -25.20) >consensus AAUUUGCCAUGCCCCAGUUUACCACGAAAUGGCCAAGCAACCCUCGUGGCCAAAAUCCAGAAGGACUGACAU_CA_G_CUUGGAGCUCC________AGGGCAUCGAAGCUGAUUAGCAU ..((((..((((((...............((((((...........)))))).....(((.....)))..............................))))))))))(((....))).. (-20.16 = -20.50 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:19 2006