
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 783,738 – 783,831 |
| Length | 93 |
| Max. P | 0.873885 |

| Location | 783,738 – 783,831 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -11.52 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
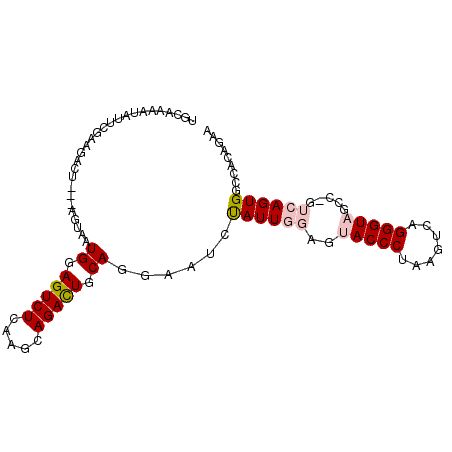
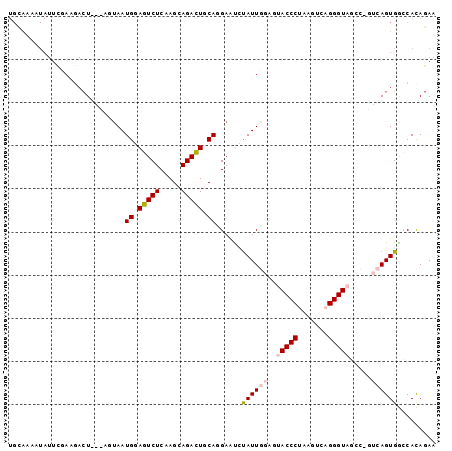
>3R_DroMel_CAF1 783738 93 + 27905053 UGCAAAGUAUUCUAAGACU---AGUAAUGCAGUCUCAAGCAGACUGCAGGAAUAUAUUGG---ACCCUAUGUCAGGGUAGCCUGUCAGUGGCCACAGAA ......(((((((......---.....((((((((.....)))))))))))))))..(((---.((...((.((((....)))).))..)))))..... ( -27.30) >DroSec_CAF1 8018 81 + 1 ------------------UAGUAGUAAUGGAGUCUCAAGCAGACUGCACGAACAUAUUGGAGUACCCAAAGUCAGGGUAGCCAGUCAGUGGCCACAGAA ------------------..((.((..((.(((((.....))))).))....((((((((..(((((.......))))).)))))..))))).)).... ( -21.20) >DroSim_CAF1 8397 99 + 1 UGUAAAAUAUUCUAGGACUAGCACUAAUGGAGUCUCAAGCAGACUGCACGAACCUAUUGGAGUACCCAAAGUCAGGGUAGCCAGUCAGUGGUCACAGAA .........((((..(((((.......((.(((((.....))))).)).....(((((((..(((((.......))))).))))).)))))))..)))) ( -26.50) >DroEre_CAF1 7314 89 + 1 UGCAAAAUAUUUGAAGACU---AUUAAUGGAGUCUCGAGCAGAUUGCAGGAAUCCAUUCGAGUACCCUAAGUCAGGGUA-------AGUGACCAUAGAA (((((..(.(((((.((((---........))))))))).)..))))).........(((..((((((.....))))))-------..)))........ ( -21.90) >DroYak_CAF1 8598 89 + 1 UGCAAAAUAUUUGAAGACU---UUAAAUGGAGUCUCAAGCAGACUUCAGGAGUCUAUUUGAGAACCCUAAGUCAGGGUA-------AGUGAGCAUAGAA (((...........(((((---(....((((((((.....)))))))).))))))........(((((.....))))).-------.....)))..... ( -24.20) >consensus UGCAAAAUAUUCGAAGACU___AGUAAUGGAGUCUCAAGCAGACUGCAGGAAUCUAUUGGAGUACCCUAAGUCAGGGUAGCC_GUCAGUGGCCACAGAA ...........................((.(((((.....))))).))......((((((..(((((.......))))).....))))))......... (-11.52 = -12.40 + 0.88)

| Location | 783,738 – 783,831 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 73.20 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -9.02 |
| Energy contribution | -9.62 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
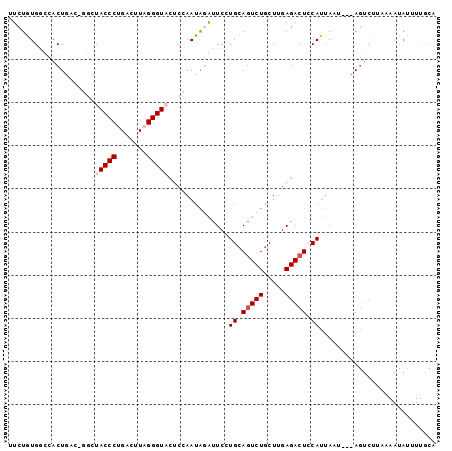
>3R_DroMel_CAF1 783738 93 - 27905053 UUCUGUGGCCACUGACAGGCUACCCUGACAUAGGGU---CCAAUAUAUUCCUGCAGUCUGCUUGAGACUGCAUUACU---AGUCUUAGAAUACUUUGCA ((((((((((.......))))))...(((..((((.---.........))))(((((((.....)))))))......---.)))..))))......... ( -25.80) >DroSec_CAF1 8018 81 - 1 UUCUGUGGCCACUGACUGGCUACCCUGACUUUGGGUACUCCAAUAUGUUCGUGCAGUCUGCUUGAGACUCCAUUACUACUA------------------ ....(((((((.....)))))))...(((.((((.....))))...))).(((.(((((.....))))).)))........------------------ ( -18.90) >DroSim_CAF1 8397 99 - 1 UUCUGUGACCACUGACUGGCUACCCUGACUUUGGGUACUCCAAUAGGUUCGUGCAGUCUGCUUGAGACUCCAUUAGUGCUAGUCCUAGAAUAUUUUACA (((((.(((((((((.(((.(((((.......)))))..))).........((.(((((.....))))).))))))))...))).)))))......... ( -25.40) >DroEre_CAF1 7314 89 - 1 UUCUAUGGUCACU-------UACCCUGACUUAGGGUACUCGAAUGGAUUCCUGCAAUCUGCUCGAGACUCCAUUAAU---AGUCUUCAAAUAUUUUGCA ....((((.....-------(((((((...)))))))(((((..(((((.....)))))..)))))...))))....---................... ( -20.40) >DroYak_CAF1 8598 89 - 1 UUCUAUGCUCACU-------UACCCUGACUUAGGGUUCUCAAAUAGACUCCUGAAGUCUGCUUGAGACUCCAUUUAA---AGUCUUCAAAUAUUUUGCA .....(((.....-------......(((((.((..((((((.((((((.....)))))).))))))..)).....)---))))............))) ( -21.50) >consensus UUCUGUGGCCACUGAC_GGCUACCCUGACUUAGGGUACUCCAAUAGAUUCCUGCAGUCUGCUUGAGACUCCAUUAAU___AGUCUUAAAAUAUUUUGCA ....................(((((.......)))))..............((.(((((.....))))).))........................... ( -9.02 = -9.62 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:51 2006