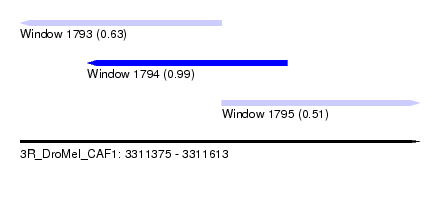
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,311,375 – 3,311,613 |
| Length | 238 |
| Max. P | 0.993825 |

| Location | 3,311,375 – 3,311,495 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -33.92 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
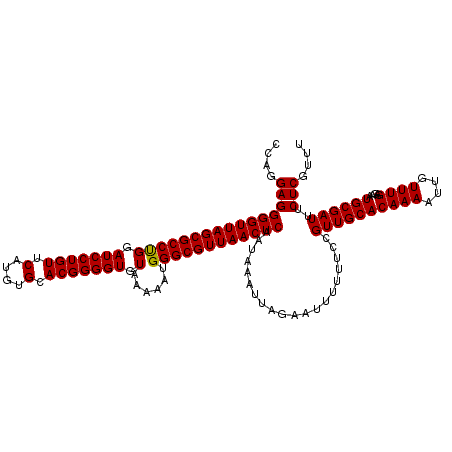
>3R_DroMel_CAF1 3311375 120 - 27905053 CCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGAGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).)......))))))))))))....................((((((((((....))))....))))))..))).... ( -33.00) >DroSec_CAF1 15559 120 - 1 CCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGAGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).......)))))))))))))....................((((((((((....))))....))))))..))).... ( -35.00) >DroSim_CAF1 19804 120 - 1 CCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGAGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).......)))))))))))))....................((((((((((....))))....))))))..))).... ( -35.00) >DroEre_CAF1 10159 120 - 1 CCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGGGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).)......)))))))))))).........(((((.((((((..(((......))).))))))...)))))))).... ( -35.20) >DroYak_CAF1 25846 120 - 1 CCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGAGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).)......))))))))))))....................((((((((((....))))....))))))..))).... ( -33.00) >consensus CCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUUCCGUUGCACAAAAUUGUUUGAGAAUGCGAUUUCUCGUUU ....((((((((((((((((.(((((((.(....).))))))).)......))))))))))))....................((((((((((....))))....))))))..))).... (-33.92 = -33.68 + -0.24)


| Location | 3,311,415 – 3,311,534 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.12 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
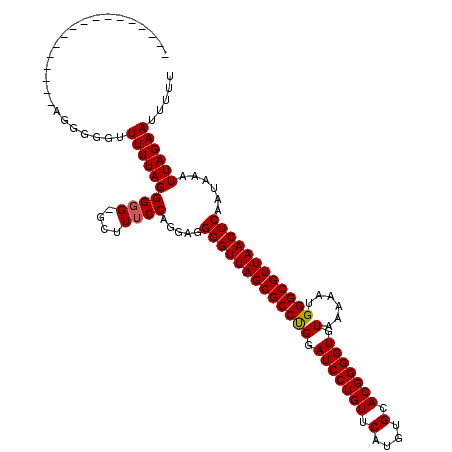
>3R_DroMel_CAF1 3311415 119 - 27905053 GAUUUUAAGGGGUUCUGAGGGGGGUUUUAGGGUG-GCUUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU .......(((((((((((((..((((.......)-)))..)).....(((((((((((((.(((((((.(....).))))))).)......))))))))))))......))))))))))) ( -41.70) >DroSec_CAF1 15599 103 - 1 -----------------AGGGGAUUUUUAGGGGGUGCUUUCCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU -----------------..((((.((((((((((....)))).....(((((((((((((.(((((((.(....).))))))).......)))))))))))))......)))))).)))) ( -32.20) >DroSim_CAF1 19844 103 - 1 -----------------AGGGGAUUUUUAGGGGGUGCUUUCCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU -----------------..((((.((((((((((....)))).....(((((((((((((.(((((((.(....).))))))).......)))))))))))))......)))))).)))) ( -32.20) >DroEre_CAF1 10199 97 - 1 ---------------------GGUUUUUAGGGGG--UGUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU ---------------------...((((((..((--....)).....(((((((((((((.(((((((.(....).))))))).)......))))))))))))......))))))..... ( -27.90) >DroYak_CAF1 25886 97 - 1 ---------------------GGUUUUUAGGGGG--CCUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU ---------------------(((((.....)))--)).........(((((((((((((.(((((((.(....).))))))).)......))))))))))))................. ( -29.00) >consensus _________________AGGGGGUUUUUAGGGGG_GCUUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUU ........................((((((((((....)))).....(((((((((((((.(((((((.(....).))))))).)......))))))))))))......))))))..... (-29.16 = -29.12 + -0.04)

| Location | 3,311,495 – 3,311,613 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
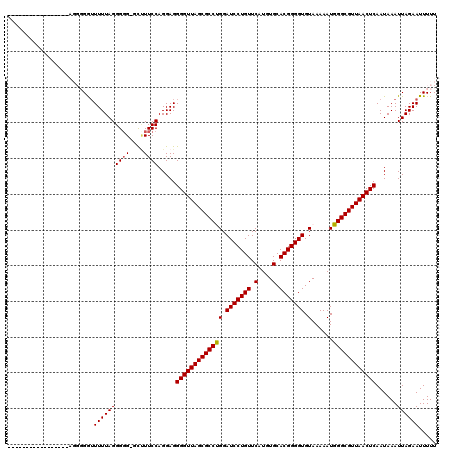
>3R_DroMel_CAF1 3311495 118 + 27905053 AAAGC-CACCCUAAAACCCCCCUCAGAACCCCUUAAAAUCCCCCAUCCGGUGGUCAUGCCCGGCCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAAAA-AAAUUGUAU .....-...........................................((((((((((..(((.....)))...).)))))))))..(((((.......))))).....-......... ( -21.50) >DroSec_CAF1 15679 103 + 1 AAAGCACCCCCUAAAAAUCCCCU-----------------CCUCCUCCGGUGGUCACGCCCGUGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAAAAAAAAUUGUAU ...(((.................-----------------.........(((((((.((..(((.(....)))).))..)))))))..(((((.......)))))..........))).. ( -19.10) >DroSim_CAF1 19924 103 + 1 AAAGCACCCCCUAAAAAUCCCCU-----------------CCCCCUCCGGUGGUCACGCCCGUGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAAAAAAAAUUGUAU ...(((.................-----------------.........(((((((.((..(((.(....)))).))..)))))))..(((((.......)))))..........))).. ( -19.10) >DroEre_CAF1 10279 93 + 1 AACA--CCCCCUAAAAACC----------------------CCCAUCCGUUGGUCACGCCCG-GCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUACAAA--AAAUUGUAU .(((--.............----------------------.......((((((((.((..(-((....)))...))..))))))..))((((.......)))).....--....))).. ( -15.70) >DroYak_CAF1 25966 94 + 1 AAGG--CCCCCUAAAAACC----------------------CCCAUCCGUUGGUCACGCCCGGGCGCAUGCCGCAGCCAUGACCAUAACACAGUCUAAUUCUGUACAAA--AAAUUCUAU .(((--...))).......----------------------.......((((((((.((...(((....)))...))..))))))..))((((.......)))).....--......... ( -15.80) >consensus AAAGC_CCCCCUAAAAACCCCCU_________________CCCCAUCCGGUGGUCACGCCCGGGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAAAA_AAAUUGUAU .................................................((((((((((....)))...((....))..)))))))..(((((.......)))))............... (-12.36 = -13.20 + 0.84)

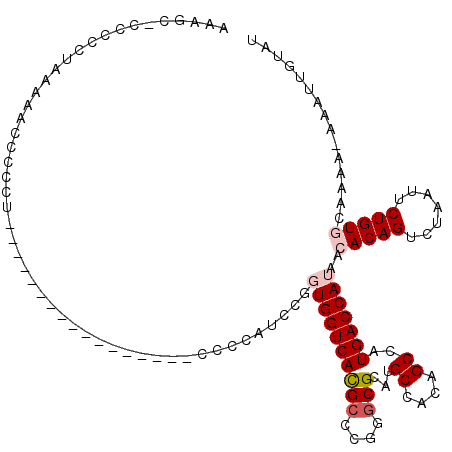
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:48 2006