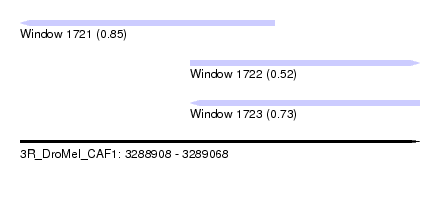
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,288,908 – 3,289,068 |
| Length | 160 |
| Max. P | 0.849120 |

| Location | 3,288,908 – 3,289,010 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -17.55 |
| Consensus MFE | -13.15 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3288908 102 - 27905053 UUUUAACGAGCUGCCAUUUGCCUCCCUUUUUCCCGCCAUAUCUCUUCGUAUAUAUCCAUAUCCGUGGCACAUAUGCAAGUUUAUAUGCCACUUACA------------CACACA .......((((........).)))..........(.(((((..(((.((((((..((((....))))...)))))))))...))))).).......------------...... ( -13.30) >DroSec_CAF1 41857 114 - 1 UUUUAACGAGCUGGCAUUUGCCUCCCUUUUUCCCGCCAUAUCUCUCCGUAUAUAUCCAUAUCCGUGCCACAUAUGCAAGCUUAUAUGCCACUUACACGCACACACUCACACACA .......(((..(((....)))............((...........((((((...(((....)))....))))))..((......)).........)).....)))....... ( -13.60) >DroEre_CAF1 43594 106 - 1 UUUUAACGAGCUGCCAUUCGCCUCCCUUUUUCCCGCCAUAUCUCUCUGUGUAUAUCCAUAUCCGUGGCACAUAUGCAAGUUUAUAUGGCACUUCC--------ACACACUCGCA ......((((..((.....)).............(((((((..((.(((((((..((((....))))...)))))))))...)))))))......--------.....)))).. ( -21.40) >DroYak_CAF1 61276 114 - 1 UUUUAACGAGCUGCCAUUCGCCUCCCUUUUUCCCGCCGUAUCUCUUUGUAUAUAUCCAUAUCCGUGGCACAUAUGCAAGUUUAUAUGGCACUUACCCACACACACACACUCACA .......(((..((.....)).............(((((((...(((((((((..((((....))))...)))))))))...)))))))...................)))... ( -21.90) >consensus UUUUAACGAGCUGCCAUUCGCCUCCCUUUUUCCCGCCAUAUCUCUCCGUAUAUAUCCAUAUCCGUGGCACAUAUGCAAGUUUAUAUGCCACUUACA_______ACACACACACA .......((((........).)))..........(((((((...(((((((((..((((....))))...)))))))))...)))))))......................... (-13.15 = -14.53 + 1.38)


| Location | 3,288,976 – 3,289,068 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3288976 92 + 27905053 GGGAAAAAGGGAGGCAAAUGGCAGCUCGUUAAAAGGAGAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUCC .........((((((...((((..(((........))).)))).))......((((.((((.....)))).)))).............)))) ( -22.00) >DroSec_CAF1 41937 92 + 1 GGGAAAAAGGGAGGCAAAUGCCAGCUCGUUAAAAGGAAAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUCC .(((....(((.(((....)))..)))....(((.((((((((((((.........))))......)))))))).)))...........))) ( -20.90) >DroSim_CAF1 43116 92 + 1 GGGAAAAGGGGAGGCAAAUGGCAGCUCGUUAAAAGGAAAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUCC .(((.((.(((..((.....))..))).)).(((.((((((((((((.........))))......)))))))).)))...........))) ( -20.50) >DroEre_CAF1 43666 92 + 1 GGGAAAAAGGGAGGCGAAUGGCAGCUCGUUAAAAGGAAAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUCC .........((((((........))((.((.(((.((((((((((((.........))))......)))))))).)))..)).))...)))) ( -20.40) >DroYak_CAF1 61356 92 + 1 GGGAAAAAGGGAGGCGAAUGGCAGCUCGUUAAAAGGAAAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUGU ........(((..((.....))..)))....(((.((((((((((((.........))))......)))))))).))).............. ( -18.70) >consensus GGGAAAAAGGGAGGCAAAUGGCAGCUCGUUAAAAGGAAAGCCGGGCGAAAUUCAAGUGCCGGAAAAUGGCUUUUGUUUAGAAGGAAAACUCC .........((((((........))((.((.(((.((((((((((((.........))))......)))))))).)))..)).))...)))) (-18.96 = -18.84 + -0.12)

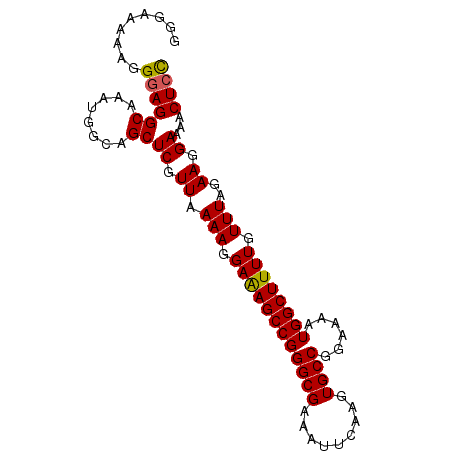
| Location | 3,288,976 – 3,289,068 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3288976 92 - 27905053 GGAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUCUCCUUUUAACGAGCUGCCAUUUGCCUCCCUUUUUCCC ((((.............(((..(((.......)))..))).........((((((..........))))))........))))......... ( -16.90) >DroSec_CAF1 41937 92 - 1 GGAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUUUCCUUUUAACGAGCUGGCAUUUGCCUCCCUUUUUCCC ((((.............(((..(((.......)))..)))......(((.(((((..........))))))))......))))......... ( -18.70) >DroSim_CAF1 43116 92 - 1 GGAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUUUCCUUUUAACGAGCUGCCAUUUGCCUCCCCUUUUCCC ((((.............(((..(((.......)))..)))..........(((.(((........)))..)))......))))......... ( -14.80) >DroEre_CAF1 43666 92 - 1 GGAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUUUCCUUUUAACGAGCUGCCAUUCGCCUCCCUUUUUCCC ((((.............(((..(((.......)))..)))..........(((.(((........)))..)))......))))......... ( -14.80) >DroYak_CAF1 61356 92 - 1 ACAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUUUCCUUUUAACGAGCUGCCAUUCGCCUCCCUUUUUCCC .((((((.......(((..((((((.......(((...........))).))))))..)))....))))))..................... ( -11.40) >consensus GGAGUUUUCCUUCUAAACAAAAGCCAUUUUCCGGCACUUGAAUUUCGCCCGGCUUUCCUUUUAACGAGCUGCCAUUUGCCUCCCUUUUUCCC ((((.............(((..(((.......)))..))).........((((((..........))))))........))))......... (-13.52 = -13.76 + 0.24)

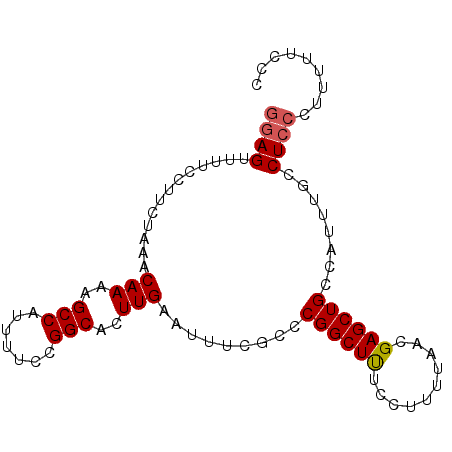
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:40 2006