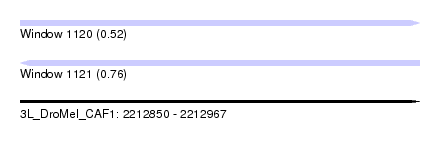
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,212,850 – 2,212,967 |
| Length | 117 |
| Max. P | 0.757132 |

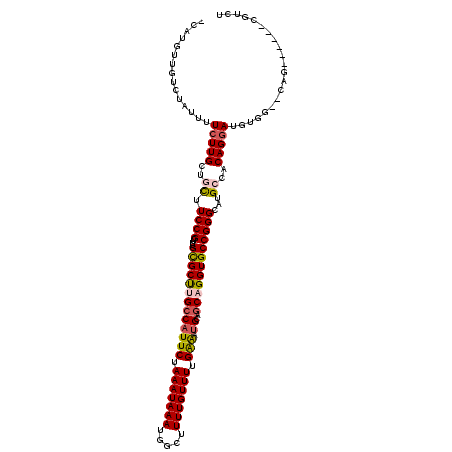
| Location | 2,212,850 – 2,212,967 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2212850 117 + 23771897 AGAGGCACAUCCUG--GCCCAUCCUGUGGCAGCCCCGGCACCUGCUGCAUCUCAAAACAAAGGCCAUUUAUUUAGAAUGGC-AGCGCAAAGCGGAAGCGCCAAGAAAAUAGACAACACCA .........((.((--((......(((((((((...(....).)))))..............(((((((.....)))))))-..))))..((....)))))).))............... ( -31.40) >DroVir_CAF1 288866 110 + 1 AGAAG------C---CACACAUCCUGUGGCAUGCCCGGCACCUGCUGCAUUUCCAAACAAAAGCCAUUUAUUUAGAAUGGCAAGCACAAAGCGGAAGCAGCAAGAAAAUAGGCAACAUG- ....(------(---((((.....)))))).((((.......((((((..((((........(((((((.....)))))))..((.....))))))))))))........)))).....- ( -38.76) >DroPse_CAF1 312392 113 + 1 UGUCGUACAUCCUG-------UCCUGUGGCAGGCCCGGCACCUGCUGCAUUCCCAAACAAAGGCCAUUUAUUUAGAAUGCCAAGCGCAAAACGGAAGCGCCAAGAAAACAGACUACAACA .((.(((....(((-------(..((..(((((.......)))))..))(((.........(((.((((.....)))))))..((((.........))))...))).))))..))).)). ( -27.80) >DroGri_CAF1 257364 111 + 1 CGACG------CUG--CCACAUCCUGUGGCAUACCCGGCACCUGCUGCGCCUCGAAACAAAAGCCAUUUAUUUAGAAUGGCAAGCACAAAGCGGAAACACCAAGAAAAUAGACAACAUG- (((.(------(((--((((.....))))))....((((....)))).)).)))........(((((((.....)))))))..((.....))(....).....................- ( -30.30) >DroWil_CAF1 340660 111 + 1 AGAGG------CUG--CUACAUCCUGUGGCAUACCCGGCACCUGCUGCAUUUCAAAACAAAAGGCAUUUAUUUAGAAUGGCGAGCGCAAAACGGAAGUAGCAAGAAAAUAGACAACAAU- .....------.((--((((.(((.((.((.....((((....))))...............(.(((((.....))))).)....))...))))).)))))).................- ( -24.60) >DroMoj_CAF1 276460 107 + 1 AGACG------CUGUCCCACAUCCUGUGUCAUGCCCGGCACCUGCUGCAUUUCAAAACAAAAGCCAUUUAUUUAGAAUGGCAGGCGCAAAACGGAAGCAGCAACAACGUAGAC------- ..(((------.(((..........(((((......))))).((((((.((((.........(((((((.....))))))).(........)))))))))))))).)))....------- ( -29.40) >consensus AGACG______CUG__CCACAUCCUGUGGCAUGCCCGGCACCUGCUGCAUUUCAAAACAAAAGCCAUUUAUUUAGAAUGGCAAGCGCAAAACGGAAGCACCAAGAAAAUAGACAACAAC_ .....................(((((((.(.....((((....))))...............(((((((.....)))))))..)))))....)))......................... (-15.15 = -15.27 + 0.11)

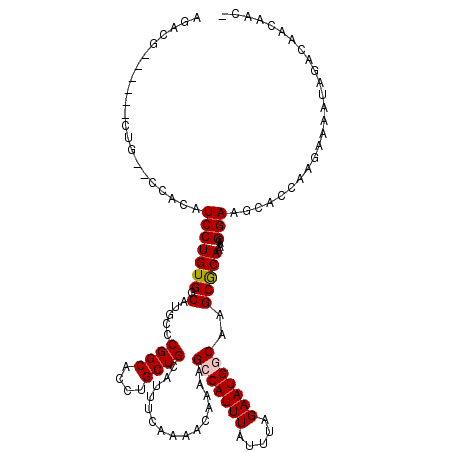
| Location | 2,212,850 – 2,212,967 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2212850 117 - 23771897 UGGUGUUGUCUAUUUUCUUGGCGCUUCCGCUUUGCGCU-GCCAUUCUAAAUAAAUGGCCUUUGUUUUGAGAUGCAGCAGGUGCCGGGGCUGCCACAGGAUGGGC--CAGGAUGUGCCUCU .((..(.........(((((((.(.(((.......(((-((..(((.(((((((.....))))))).)))..))))).(((((....)).)))...))).).))--))))).)..))... ( -38.90) >DroVir_CAF1 288866 110 - 1 -CAUGUUGCCUAUUUUCUUGCUGCUUCCGCUUUGUGCUUGCCAUUCUAAAUAAAUGGCUUUUGUUUGGAAAUGCAGCAGGUGCCGGGCAUGCCACAGGAUGUGUG---G------CUUCU -.....(((((.....((((((((((((((.....))..((((((.......))))))........))))..))))))))....))))).((((((.....))))---)------).... ( -41.00) >DroPse_CAF1 312392 113 - 1 UGUUGUAGUCUGUUUUCUUGGCGCUUCCGUUUUGCGCUUGGCAUUCUAAAUAAAUGGCCUUUGUUUGGGAAUGCAGCAGGUGCCGGGCCUGCCACAGGA-------CAGGAUGUACGACA .((((((.((((((((..(((((..((((....(((((((((((((((((((((.....))))))).)))))))..)))))))))))..))))).))))-------))))...)))))). ( -44.50) >DroGri_CAF1 257364 111 - 1 -CAUGUUGUCUAUUUUCUUGGUGUUUCCGCUUUGUGCUUGCCAUUCUAAAUAAAUGGCUUUUGUUUCGAGGCGCAGCAGGUGCCGGGUAUGCCACAGGAUGUGG--CAG------CGUCG -.((((((.((((.((((((((((.((((...(.((((((((..((.(((((((.....))))))).)))))).)))).)...)))).)))))).)))).))))--)))------))).. ( -37.90) >DroWil_CAF1 340660 111 - 1 -AUUGUUGUCUAUUUUCUUGCUACUUCCGUUUUGCGCUCGCCAUUCUAAAUAAAUGCCUUUUGUUUUGAAAUGCAGCAGGUGCCGGGUAUGCCACAGGAUGUAG--CAG------CCUCU -................(((((((.(((.....((((..(((((((.(((((((.....))))))).).))))..))..))))..((....))...))).))))--)))------..... ( -28.20) >DroMoj_CAF1 276460 107 - 1 -------GUCUACGUUGUUGCUGCUUCCGUUUUGCGCCUGCCAUUCUAAAUAAAUGGCUUUUGUUUUGAAAUGCAGCAGGUGCCGGGCAUGACACAGGAUGUGGGACAG------CGUCU -------....((((((((.(..(.((((((..(((((((((((((.(((((((.....))))))).).))))..))))))))..))).((...))))).)..))))))------))).. ( -37.30) >consensus _CAUGUUGUCUAUUUUCUUGCUGCUUCCGCUUUGCGCUUGCCAUUCUAAAUAAAUGGCUUUUGUUUUGAAAUGCAGCAGGUGCCGGGCAUGCCACAGGAUGUGG__CAG______CGUCU ...............(((((..((.((((....(((((((((((((.(((((((.....))))))).))).))..))))))))))))...))..)))))..................... (-20.74 = -21.05 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:41 2006