
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,111,296 – 2,111,444 |
| Length | 148 |
| Max. P | 0.993318 |

| Location | 2,111,296 – 2,111,404 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -38.09 |
| Consensus MFE | -17.22 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
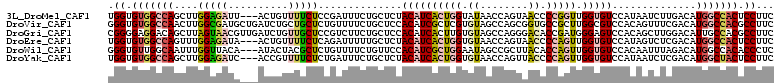
>3L_DroMel_CAF1 2111296 108 + 23771897 UGGUGUGGCCAGCUUGGAGAUU---ACUGUUUUCUCCGAUUUCUGCUCUACAUCACUGGUAUAACCAGUAACCCCGGUUGGUGUCCAUAAUCUUGACAUGGCCACUCCUUC .((.((((((((((((((((..---.......))))))).....))...((((((((((.....)))))(((....))))))))..............))))))).))... ( -36.50) >DroVir_CAF1 183739 111 + 1 GGGUGUGGCCAACUUGGCGAUGCUGAUCUGCUGCUCUGUUUUCUGCUCCACAUCGCUCGUGUAGCCAGCGGUGCCGCUUGGCGUCCACAGUUUCGACAUGGCCACGCCUUC (((((((((((....(((((((.((....((.............))..))))))))).(((..(((((((....))).))))...)))..........))))))))))).. ( -46.72) >DroGri_CAF1 158722 111 + 1 CGGGGAGGACAGCUUAGUAACGUUGAUCUGUUGCUCCGUCUUCUGCUCCACAUCACUUGUGUAGCCAGGGACACCGAUGGGAGUCCACAGCUUGGACAUUGCCACGCCUUC ((((((((((((.((((.....)))).))))).))))(((((..(((.((((.....)))).)))..))))).))).(((.((((((.....)))))..).)))....... ( -37.40) >DroEre_CAF1 167266 108 + 1 UGGUGUGGCCAGUUUGGAGAUA---ACUGUUUUCUCAGAUUUUUGCUCUACAUCACUGGUGUAACCAGUAACCCCAGUUGGUGUCCAUAGUCUCGACAUGGCCACUCCUUC .((.((((((((((..(((((.---.(((......)))...........((((((((((.((........)).)))).)))))).....)))))))).))))))).))... ( -37.40) >DroWil_CAF1 206067 108 + 1 GGGUGUUGGCAAUUUGGUUACA---AUACUACGCUCUGUUUUCUGUUCCACAUCGCUGGAAUAGCCGCUUACACCAGUUGGUGUCCACAAUUUAGACAUGGCCACACCCUC ((((((.(((....((((....---..)))).(.((((....((((((((......))))))))......(((((....)))))........)))))...))))))))).. ( -33.20) >DroYak_CAF1 161015 108 + 1 UGGUGUGGCCAGCUUGGAGAUC---ACCGUUUUCUCUGAUUUCUGCUCUACAUCACUGGUGUAACCAGUUACCCCAGUUGGUGUCCAUAAUCUCGACAUGGCUACUCCUUC .((.(((((((((..(((((((---(..(....)..))))))))))...((((((((((.((((....)))).)))).))))))..............))))))).))... ( -37.30) >consensus UGGUGUGGCCAGCUUGGAGAUA___ACCGUUUGCUCUGAUUUCUGCUCCACAUCACUGGUGUAACCAGUAACCCCAGUUGGUGUCCACAAUCUCGACAUGGCCACUCCUUC .((.(((((((....(((((..........)))))..............((((((((((.((........)).))))).)))))..............))))))).))... (-17.22 = -18.28 + 1.06)
| Location | 2,111,296 – 2,111,404 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2111296 108 - 23771897 GAAGGAGUGGCCAUGUCAAGAUUAUGGACACCAACCGGGGUUACUGGUUAUACCAGUGAUGUAGAGCAGAAAUCGGAGAAAACAGU---AAUCUCCAAGCUGGCCACACCA ...((.(((((((.((...((((.((....((....)).((((((((.....))))))))......))..))))(((((.......---..)))))..))))))))).)). ( -37.50) >DroVir_CAF1 183739 111 - 1 GAAGGCGUGGCCAUGUCGAAACUGUGGACGCCAAGCGGCACCGCUGGCUACACGAGCGAUGUGGAGCAGAAAACAGAGCAGCAGAUCAGCAUCGCCAAGUUGGCCACACCC ...((.(((((((..........(((...((((.(((....)))))))..)))(.(((((((.((((.............))...)).))))))))....))))))).)). ( -42.92) >DroGri_CAF1 158722 111 - 1 GAAGGCGUGGCAAUGUCCAAGCUGUGGACUCCCAUCGGUGUCCCUGGCUACACAAGUGAUGUGGAGCAGAAGACGGAGCAACAGAUCAACGUUACUAAGCUGUCCUCCCCG ((.(((.(((.(((((...(((((.((((.((....)).)))).))))).......((((.((..((..........))..)).))))))))).))).))).))....... ( -29.90) >DroEre_CAF1 167266 108 - 1 GAAGGAGUGGCCAUGUCGAGACUAUGGACACCAACUGGGGUUACUGGUUACACCAGUGAUGUAGAGCAAAAAUCUGAGAAAACAGU---UAUCUCCAAACUGGCCACACCA ...((.(((((((((((.(.....).))))..(((((..((((((((.....)))))))).((((.......))))......))))---)..........))))))).)). ( -36.80) >DroWil_CAF1 206067 108 - 1 GAGGGUGUGGCCAUGUCUAAAUUGUGGACACCAACUGGUGUAAGCGGCUAUUCCAGCGAUGUGGAACAGAAAACAGAGCGUAGUAU---UGUAACCAAAUUGCCAACACCC ..(((((((((((((((.....(((..(((((....)))))..)))(((.....))))))))).........((((.((...)).)---))).........))).)))))) ( -32.10) >DroYak_CAF1 161015 108 - 1 GAAGGAGUAGCCAUGUCGAGAUUAUGGACACCAACUGGGGUAACUGGUUACACCAGUGAUGUAGAGCAGAAAUCAGAGAAAACGGU---GAUCUCCAAGCUGGCCACACCA ...((.((.((((.((.((((((((.((((...(((((.((((....)))).)))))..))).((.......))........).))---))))))...)))))).)).)). ( -33.20) >consensus GAAGGAGUGGCCAUGUCGAAACUAUGGACACCAACCGGGGUAACUGGCUACACCAGUGAUGUAGAGCAGAAAACAGAGAAAACGAU___CAUCUCCAAGCUGGCCACACCA ...((.(((((((((((.........))))......(((((((((((.....))))))))(.....)..........................)))....))))))).)). (-18.74 = -20.13 + 1.39)

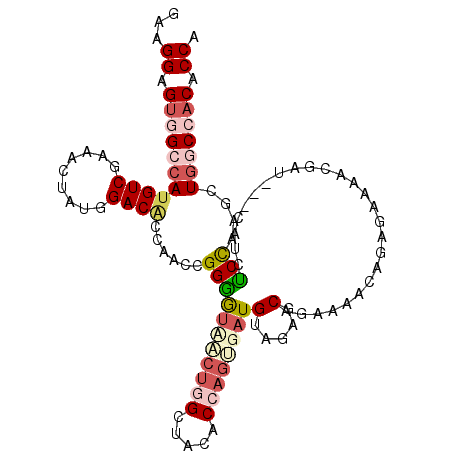
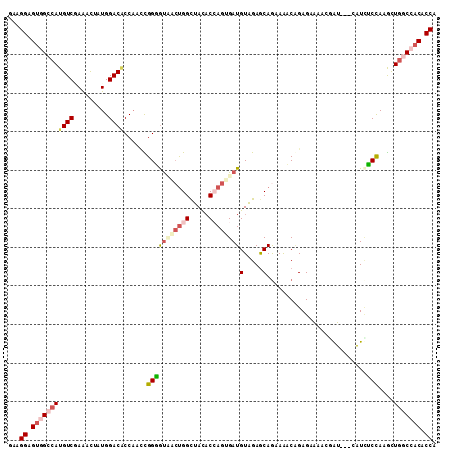
| Location | 2,111,330 – 2,111,444 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -21.77 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2111330 114 - 23771897 CCAGCAGAACCCACGCUCACCAUCUCCAAAACCCUCCGCCGAAGGAGUGGCCAUGUCAAGAUUAUGGACACCAACCGGGGUUACUGGUUAUACCAGUGAUGUAGAGCAGAAAUC ..............((((..((((.....(((((((((((....).))))(((((.......))))).........))))))(((((.....)))))))))..))))....... ( -35.10) >DroVir_CAF1 183776 114 - 1 ACAGCAAAAUACGCGCUCGCCAUCGCCGAAGCCAUCGGCUGAAGGCGUGGCCAUGUCGAAACUGUGGACGCCAAGCGGCACCGCUGGCUACACGAGCGAUGUGGAGCAGAAAAC ...((.......))((((((((.((((..((((...))))...))))))))(((((((....((((...((((.(((....)))))))..))))..)))))))))))....... ( -50.70) >DroGri_CAF1 158759 114 - 1 ACAGCAAAAUACACGUUCGCCAUCUCCAAAGCCAUCGGCGGAAGGCGUGGCAAUGUCCAAGCUGUGGACUCCCAUCGGUGUCCCUGGCUACACAAGUGAUGUGGAGCAGAAGAC ..............((((..((((.(....((((.((.(....).))))))..(((...(((((.((((.((....)).)))).)))))..))).).))))..))))....... ( -36.70) >DroEre_CAF1 167300 114 - 1 CCAGCAGAACCCACGCUCACCCUCUCCCAAGCCCUCCGCCGAAGGAGUGGCCAUGUCGAGACUAUGGACACCAACUGGGGUUACUGGUUACACCAGUGAUGUAGAGCAAAAAUC ..............((((.......((((.(((((((......)))).)))..((((.(.....).)))).....))))((((((((.....))))))))...))))....... ( -38.70) >DroWil_CAF1 206101 114 - 1 CCAGCAGAAUCCGCGCUCACCUUCACCCAAACCCUCGGCUGAGGGUGUGGCCAUGUCUAAAUUGUGGACACCAACUGGUGUAAGCGGCUAUUCCAGCGAUGUGGAACAGAAAAC .........((((((((((((((((.((........)).)))))))).)))...(((.....(((..(((((....)))))..)))(((.....)))))))))))......... ( -37.20) >DroMoj_CAF1 162638 114 - 1 CCAGCAGAACACGCGUUCGCCAUCGCCCAAGCCAUCGGCAGAGGGCGUGGCCAUGUCUAAGCUGUGGACACCUAGUGGCCCCACAAGCUACUCUAGCGAUGUGGAGCAGAAGAC .((((.((((....))))((((.(((((..(((...)))...))))))))).........))))....(((...)))((.(((((.((((...))))..))))).))....... ( -42.20) >consensus CCAGCAGAACACACGCUCACCAUCUCCCAAGCCAUCGGCCGAAGGCGUGGCCAUGUCGAAACUGUGGACACCAACCGGCGUCACUGGCUACACCAGCGAUGUGGAGCAGAAAAC ..............((((..((((......(((.((....)).)))((((((.((((.........))))((....)).......))))))......))))..))))....... (-21.77 = -22.22 + 0.45)

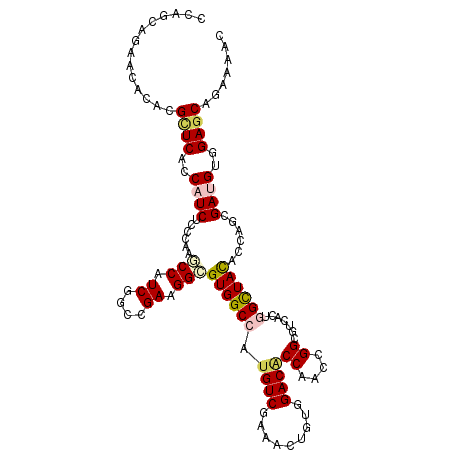
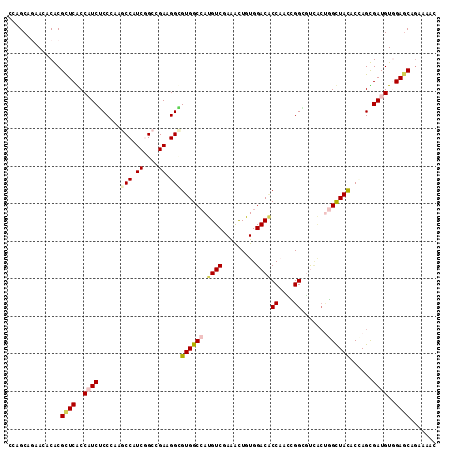
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:58 2006