| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,918,990 – 1,919,099 |
| Length | 109 |
| Max. P | 0.703468 |

| Location | 1,918,990 – 1,919,099 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.23 |
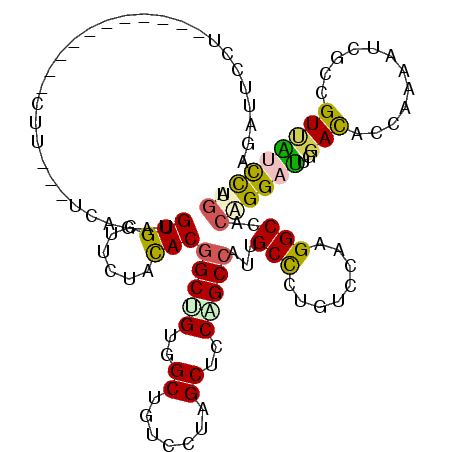
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -17.63 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.41 |
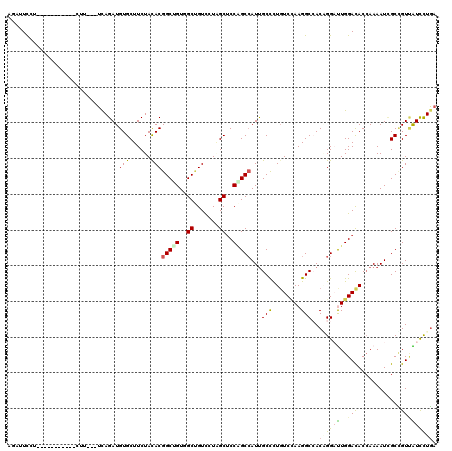
| Combinations/Pair | 1.50 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
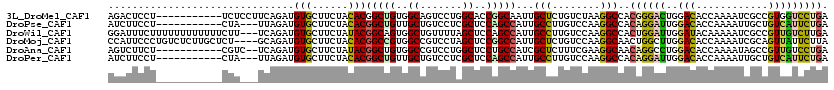
>3L_DroMel_CAF1 1918990 109 - 23771897 AGACUCCU-----------UCUCCUUCAGAUGUGCUUCUACACGGCUGUGGCAGUCCUGGCACCGGCAAUUGCUCUGUCUAAGGCCACGGGACUGGACACCAAAAUCGCCGUGGUCCUGA .(((....-----------........(((......))).((((((..(((((((((((((...((((.......))))....)))..)))))))....))).....))))))))).... ( -33.30) >DroPse_CAF1 48696 106 - 1 AUCUUCCU-----------CUA---UUAGAUGUGCUUCUACACGGCUGUUGCUGUCCUCGCUCCAGCCAUUGCCUUGUCCAAGGCCACAGGAUUGGACACCAAAAUUGCUGUCAUUCUGA ((((....-----------...---..))))(((......)))(((((..((.......))..)))))...(((((....)))))..((((((..((((.((....)).)))))))))). ( -28.50) >DroWil_CAF1 63296 117 - 1 GGAUUUCUUUUUUUUUUUUCUU---UCAGAUGUGCUUCUAUACGGCAGUGGCUGUUUUAGCUCCAGCCAUUGCCUUGUCCAAGGCCACUGGAUUGGAUACAAAAAUCGCCGUUGUCUUGA ((((..(....(((((..((((---((((.((.((((..(((.(((((((((((.........))))))))))).)))...)))))))))))..)))...))))).....)..))))... ( -32.70) >DroMoj_CAF1 52097 116 - 1 CCAUUCCCUGUCUCUUGCUCU----GCAGAUGUGCUUCUACACGGCCGUGGCCGUCCUAGCUCCGGCCAUUGCUCUGUCCAAGGCAACUGGCUUGGACACCAAAAUCGCAGUUAUUCUUA ..............((((...----((((..(((......)))(((((..((.......))..))))).))))..((((((((.(....).))))))))........))))......... ( -36.20) >DroAna_CAF1 45343 107 - 1 AGUCUUCU-----------CGUC--UCAGAUGUGCUUCUAUACGGCUGUGGCCGUCCUGGCUCCUGCCAUCGCUCUUUCGAAGGCAACAGGCCUGGACACCAAAAUAGCCGUUGUCCUGA ........-----------....--((((....((......((((((((((..((((.((((..((((.(((......))).))))...)))).)))).)))...))))))).)).)))) ( -34.80) >DroPer_CAF1 47929 106 - 1 AUCUUCCU-----------CUA---UUAGAUGUGCUUCUACACGGCUGUUGCUGUCCUCGCUCCAGCCAUUGCCUUGUCCAAGGCCACAGGAUUGGACACCAAAAUUGCUGUCAUUCUGA ((((....-----------...---..))))(((......)))(((((..((.......))..)))))...(((((....)))))..((((((..((((.((....)).)))))))))). ( -28.50) >consensus AGAUUCCU___________CUU___UCAGAUGUGCUUCUACACGGCUGUGGCUGUCCUAGCUCCAGCCAUUGCCCUGUCCAAGGCCACAGGAUUGGACACCAAAAUCGCCGUUAUCCUGA ...............................(((......)))(((((..((.......))..)))))...(((........)))..((((((..(((............))))))))). (-17.63 = -17.22 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:32 2006