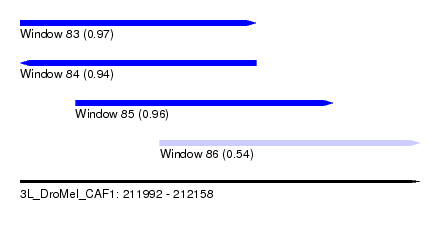
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 211,992 – 212,158 |
| Length | 166 |
| Max. P | 0.970645 |

| Location | 211,992 – 212,090 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -40.29 |
| Consensus MFE | -28.45 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
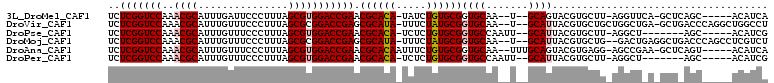
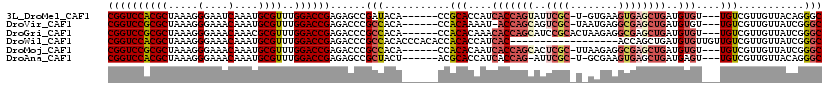
>3L_DroMel_CAF1 211992 98 + 23771897 UGAUGU-----GCUGAGC-UGAACCU-AAGCACGUACUGC--A--UUGCACCGCACAGAUA-UGUGCGUUCGGUCCACGCUAAAGGGAAUCAAAUGCGUUUGGACCGAGA ..((((-----(((.((.-.....))-.))))))).....--.--......((((((....-))))))((((((((((((...............))))..)))))))). ( -34.36) >DroVir_CAF1 4398 104 + 1 AGGCCAGCCUGGGUCAGC-UCAGCCAGCAGCACGUAAUGC--A--UUGCACCGCAUAGAAA-UAUGCGCUCGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA .(((.(((........))-)..))).((((((.....)))--.--.)))..((((((....-))))))((((((((((((.....(....)....))))..)))))))). ( -42.20) >DroPse_CAF1 4859 94 + 1 CGAUGU-----GCU-------AGCCU-AAGCACGUAAUGC--AAUUGGCACCGCACAGAGA-UGUGCGUUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA ..((((-----(((-------.....-.)))))))..(((--.....))).((((((....-))))))((((((((((((.....(....)....))))..)))))))). ( -39.00) >DroMoj_CAF1 5468 103 + 1 AGACGAGGCUGGGUCAGCCUCAGUC--CAGCACGUAAUGC--A--UUGCACCGCAUAGAAA-UAUGCGCUCGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA .((((((((((...))))))).)))--..(((.....)))--.--......((((((....-))))))((((((((((((.....(....)....))))..)))))))). ( -47.40) >DroAna_CAF1 6072 101 + 1 UGAUGU-----ACUGAGC-UUCGGCU-CCUCACGUACUGCAAA--UUGCACCGCACAGAAAUUGUGCGUUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA ..((((-----...((((-....)))-)...))))..(((...--..))).(((((((...)))))))((((((((((((.....(....)....))))..)))))))). ( -39.80) >DroPer_CAF1 4862 94 + 1 CGAUGU-----GCU-------AGCCU-AAGCACGUAAUGC--AAUUGGCACCGCACAGAGA-UGUGCGUUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA ..((((-----(((-------.....-.)))))))..(((--.....))).((((((....-))))))((((((((((((.....(....)....))))..)))))))). ( -39.00) >consensus AGAUGU_____GCU_AGC_U_AGCCU_AAGCACGUAAUGC__A__UUGCACCGCACAGAAA_UGUGCGUUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGA .....................................(((.......))).((((((.....))))))((((((((((((.....(....)....))))..)))))))). (-28.45 = -27.57 + -0.89)

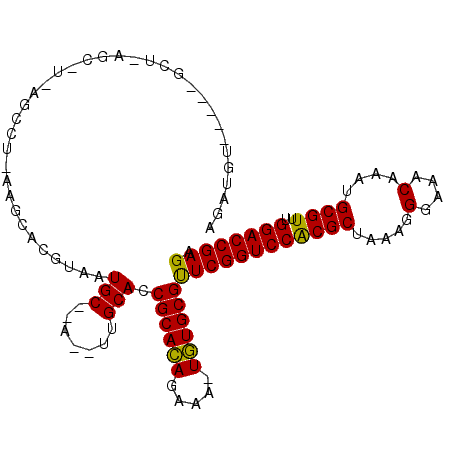
| Location | 211,992 – 212,090 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.09 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 211992 98 - 23771897 UCUCGGUCCAAACGCAUUUGAUUCCCUUUAGCGUGGACCGAACGCACA-UAUCUGUGCGGUGCAA--U--GCAGUACGUGCUU-AGGUUCA-GCUCAGC-----ACAUCA ..(((((((..((((...............))))))))))).((((((-....))))))((((..--.--...))))(((((.-((.....-.)).)))-----)).... ( -36.06) >DroVir_CAF1 4398 104 - 1 UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGCGGACCGAGCGCAUA-UUUCUAUGCGGUGCAA--U--GCAUUACGUGCUGCUGGCUGA-GCUGACCCAGGCUGGCCU .((((((((...(((...............))).))))))))((((((-....))))))..(((.--.--((((...))))))).((((.(-(((......)))))))). ( -37.06) >DroPse_CAF1 4859 94 - 1 UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGUGGACCGAACGCACA-UCUCUGUGCGGUGCCAAUU--GCAUUACGUGCUU-AGGCU-------AGC-----ACAUCG ..(((((((..((((...............))))))))))).((((((-....))))))((((.....--))))...(((((.-.....-------)))-----)).... ( -33.66) >DroMoj_CAF1 5468 103 - 1 UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGCGGACCGAGCGCAUA-UUUCUAUGCGGUGCAA--U--GCAUUACGUGCUG--GACUGAGGCUGACCCAGCCUCGUCU .((((((((...(((...............))).))))))))((((((-....))))))......--.--((((...)))).(--(((.(((((((...))))))))))) ( -39.36) >DroAna_CAF1 6072 101 - 1 UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGUGGACCGAACGCACAAUUUCUGUGCGGUGCAA--UUUGCAGUACGUGAGG-AGCCGAA-GCUCAGU-----ACAUCA ..(((((((..((((...............))))))))))).((((((.....))))))((((..--......))))(((..(-(((....-))))..)-----)).... ( -36.56) >DroPer_CAF1 4862 94 - 1 UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGUGGACCGAACGCACA-UCUCUGUGCGGUGCCAAUU--GCAUUACGUGCUU-AGGCU-------AGC-----ACAUCG ..(((((((..((((...............))))))))))).((((((-....))))))((((.....--))))...(((((.-.....-------)))-----)).... ( -33.66) >consensus UCUCGGUCCAAACGCAUUUGUUUCCCUUUAGCGUGGACCGAACGCACA_UUUCUGUGCGGUGCAA__U__GCAUUACGUGCUG_AGGCU_A_GCU_AGC_____ACAUCA ..(((((((..((((...............))))))))))).((((((.....))))))((((.......)))).................................... (-23.87 = -24.09 + 0.22)
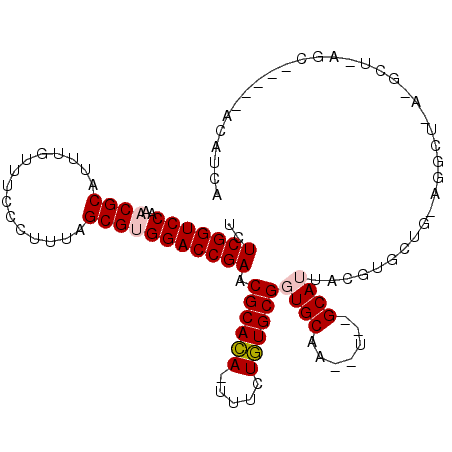
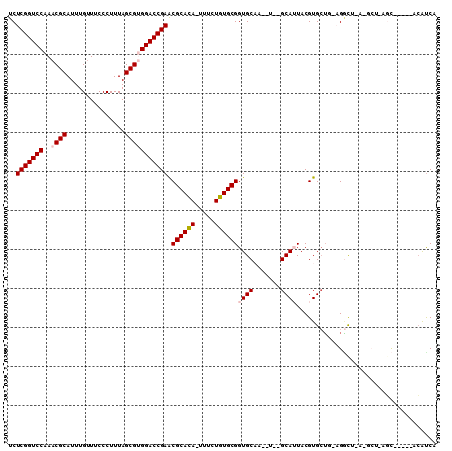
| Location | 212,015 – 212,122 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -29.45 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
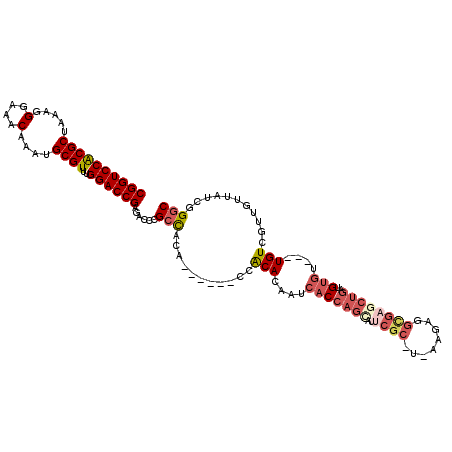
>3L_DroMel_CAF1 212015 107 + 23771897 CACGUACUGC--AUUGCACCGCACAGAUA-UGUGCGUUCGGUCCACGCUAAAGGGAAUCAAAUGCGUUUGGACCGAGAGCCGAUACA------CCGCACCAUCACCAGUAUUCGC-U- ...((((((.--..(((..((((((....-))))))((((((((((((...............))))..))))))))..........------..))).......))))))....-.- ( -32.16) >DroVir_CAF1 4427 107 + 1 CACGUAAUGC--AUUGCACCGCAUAGAAA-UAUGCGCUCGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA------CCACAAAAU-ACCAGCAGUCGC-UA ........((--(((((..((((((....-))))))((((((((((((.....(....)....))))..))))))))..........------.........-....))))).))-.. ( -39.30) >DroGri_CAF1 619 109 + 1 CACGUAAUGC--AUUGCACCGCAUAGAAA-UAUGCGCUCGGUCCGCGCUAAAGGGAAACAAACGCGUUUGGACCGAGACCCGCCACA------CCACACAAACACCAGCAUCCGCACU .......(((--..(((..((((((....-))))))((((((((((((.....(....)....))))..))))))))..........------..............)))...))).. ( -37.20) >DroWil_CAF1 5075 104 + 1 CGUAUAAUGCACAUUGCACCGCACAGAAA-UGUGCGCUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACACCCACACCACACCAUCAC------------- .......(((.....))).((((((....-))))))((((((((((((.....(....)....))))..))))))))............................------------- ( -35.20) >DroMoj_CAF1 5496 108 + 1 CACGUAAUGC--AUUGCACCGCAUAGAAA-UAUGCGCUCGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA------CCACACAAUCACCAGCACUCGC-UU ........((--..(((..((((((....-))))))((((((((((((.....(....)....))))..))))))))..........------..............)))...))-.. ( -36.00) >DroAna_CAF1 6095 109 + 1 CACGUACUGCAAAUUGCACCGCACAGAAAUUGUGCGUUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGAGCCGCUACU------ACGCACCAUCACCAG-AUUCGC-U- ........((.((((((..(((((((...)))))))((((((((((((.....(....)....))))..)))))))).)).((....------..))..........)-))).))-.- ( -33.60) >consensus CACGUAAUGC__AUUGCACCGCACAGAAA_UAUGCGCUCGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA______CCACACAAUCACCAGCAUUCGC_U_ .......(((.....))).((((((.....))))))((((((((((((.....(....)....))))..))))))))......................................... (-29.45 = -28.48 + -0.97)
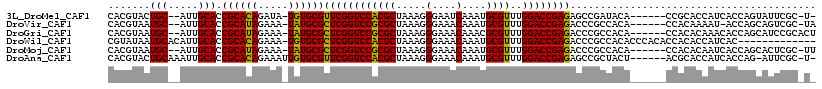
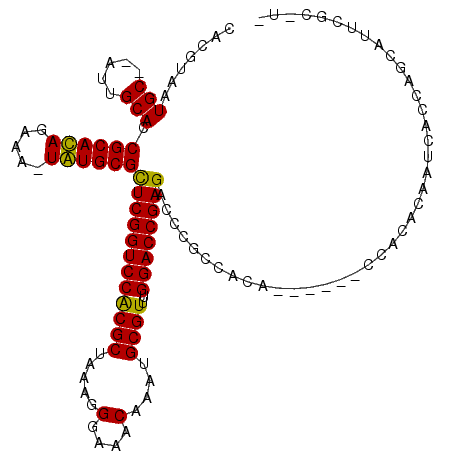
| Location | 212,050 – 212,158 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -37.49 |
| Consensus MFE | -23.98 |
| Energy contribution | -25.20 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 212050 108 + 23771897 CGGUCCACGCUAAAGGGAAUCAAAUGCGUUUGGACCGAGAGCCGAUACA------CCGCACCAUCACCAGUAUUCGC-U-GUGAAGUGAGCUGAUGUGU---UGUCGUUGUUACAGGGC ((((((((((...............))))..))))))...(((...(((------.((((.((.((.((((..((((-(-....))))))))).)))).---)).)).))).....))) ( -32.86) >DroVir_CAF1 4462 108 + 1 CGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA------CCACAAAAU-ACCAGCAGUCGC-UAAUGAGGCGAGCUGAUGUGU---UGUCGUUGUUAUCGGGC ((((((((((.....(....)....))))..))))))...((((..(((------(.((((.((-(.((((..((((-(.....))))))))).))).)---))).).)))...)))). ( -41.20) >DroGri_CAF1 654 110 + 1 CGGUCCGCGCUAAAGGGAAACAAACGCGUUUGGACCGAGACCCGCCACA------CCACACAAACACCAGCAUCCGCACUAAGAGGCGAGCUGAUGUGU---UGUCGUUGUUAUCGGGC ((((((((((.....(....)....))))..))))))...((((....(------(.(((((((((.((((...(((........))).)))).))).)---))).)).))...)))). ( -37.80) >DroWil_CAF1 5112 101 + 1 CGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACACCCACACCACACCAUCAC------------------ACCAGCUGAUGUGUUGUUGUCGUUGUUAUCGGGC ((((((((((.....(....)....))))..))))))...((((..(((.(.((((.((((.((((.------------------......)))))))).).))).).)))...)))). ( -33.40) >DroMoj_CAF1 5531 109 + 1 CGGUCCGCGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA------CCACACAAUCACCAGCACUCGC-UUAAGAGGCGAGCUGAUGUGU---UGUCGUUGUUAUCGGGC ((((((((((.....(....)....))))..))))))...((((....(------(.(((((((((.((((..((((-(.....))))))))).)).))---))).)).))...)))). ( -41.40) >DroAna_CAF1 6133 107 + 1 CGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGAGCCGCUACU------ACGCACCAUCACCAG-AUUCGC-U-GCGAAGUGAGCUGAUGAGU---UGUCGUUGUUACAGGGC ((((((((((.....(....)....))))..))))))...(((.((((.------(((((...(((.(((-.(((((-(-....))))))))).)))..---)).))).))...))))) ( -38.30) >consensus CGGUCCACGCUAAAGGGAAACAAAUGCGUUUGGACCGAGACCCGCCACA______CCACACAAUCACCAGCAUUCGC_U_AAGAGGCGAGCUGAUGUGU___UGUCGUUGUUAUCGGGC ((((((((((.....(....)....))))..))))))......(((...........(((....(((((((..((((........))))))))..)))....)))...........))) (-23.98 = -25.20 + 1.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:19 2006