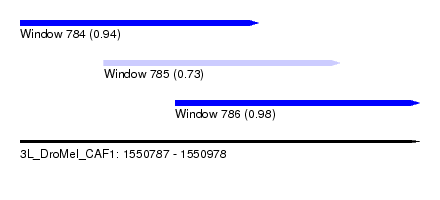
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,550,787 – 1,550,978 |
| Length | 191 |
| Max. P | 0.980556 |

| Location | 1,550,787 – 1,550,901 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -54.12 |
| Consensus MFE | -43.86 |
| Energy contribution | -44.25 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1550787 114 + 23771897 CGGUGGUGAAUCCUCCAAACGGAGGUAUCCUGCCGCCUCU---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUAGGCAUCA .(((((.((..(((((....))))).....((((((((..---(....)))))---)))))).))))).((((.(((....)))))))(((.((((((........)))))).))).... ( -54.20) >DroPse_CAF1 10316 111 + 1 AG---------CAGCCAAGUGGUGGAAUACUGCCGCCUCUUCCGGCAGUCGGCAAUGGGAUCUCCACCAGCCAGCGGCCCUAUGUGGCGCCGAGGCUGCGCAACAAUGGCCUUGGCAUCA ..---------..((((.((((.((...(((((((.......))))))).(((..(((........)))))).....)))))).))))(((((((((..........))))))))).... ( -47.20) >DroSec_CAF1 8463 114 + 1 CGGUGGUGUCUCCUCCCAACGGAGGUAUCCUGCCGCCUCU---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCA .((..(.(...(((((....)))))...))..))(((.(.---((((...(((---(((......))).)))....))))...).)))((((((((((........)))))))))).... ( -58.10) >DroSim_CAF1 8581 114 + 1 CGGUGGUGUCUCCUCCCAACGGAGGUAUCCUGCCGCCUCU---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCA .((..(.(...(((((....)))))...))..))(((.(.---((((...(((---(((......))).)))....))))...).)))((((((((((........)))))))))).... ( -58.10) >DroYak_CAF1 9146 114 + 1 CGGUGGUGACUCCUCCGAACGGAGGUAUCCUGCCGCCUCU---GGGAACGGGC---GGAAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGGAACAACGGCCUGGGCAUCA .(((((.((.(((((((..(((((((........))))))---)....)))).---))).)).))))).((((.(((....)))))))((((((((((........)))))))))).... ( -59.90) >DroPer_CAF1 10002 111 + 1 AG---------CAGCCAAGUGGUGGAAUACUGCCGCCUCUUCCGGCAGUCGGCAAUGGGAUCUCCACCAGCCAGCGGCCCUAUGUGGCGCCGAGGCUGCGCAACAAUGGCCUUGGCAUCA ..---------..((((.((((.((...(((((((.......))))))).(((..(((........)))))).....)))))).))))(((((((((..........))))))))).... ( -47.20) >consensus CGGUGGUG_CUCCUCCAAACGGAGGUAUCCUGCCGCCUCU___GGGAACGGGC___GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCA .(((((.....(((((....)))))....)))))(((.(....(((....(((...((........)).)))....)))....).)))((((((((((........)))))))))).... (-43.86 = -44.25 + 0.39)

| Location | 1,550,827 – 1,550,940 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -48.64 |
| Consensus MFE | -35.21 |
| Energy contribution | -36.68 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1550827 113 + 23771897 ---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUAGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAU-UCAGAUUCCCG ---((((((((((---(((......)))..((.(.((.....((.(((((((((((((........))))))..((....)).)))))))))..)).).)))))))..-.....))))). ( -48.31) >DroPse_CAF1 10347 102 + 1 UCCGGCAGUCGGCAAUGGGAUCUCCACCAGCCAGCGGCCCUAUGUGGCGCCGAGGCUGCGCAACAAUGGCCUUGGCAUCAGUCGGGGAC---------------CGGUGUCACCUUU--- ....(((((((((..(((........))))))..((((((.....)).)))).))))))...(((.((((((((((....))))))).)---------------)).))).......--- ( -41.80) >DroSec_CAF1 8503 113 + 1 ---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAU-UCAGAUUCCCG ---((((((((((---(((......))).((..(((.((((...((..((((((((((........))))))))))..))..)))))))...)).......)))))..-.....))))). ( -54.41) >DroSim_CAF1 8621 113 + 1 ---GGGAACGGGC---GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAU-UCAGAUUCCCG ---((((((((((---(((......))).((..(((.((((...((..((((((((((........))))))))))..))..)))))))...)).......)))))..-.....))))). ( -54.41) >DroYak_CAF1 9186 113 + 1 ---GGGAACGGGC---GGAAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGGAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGUCCGAU-UCAGAUUCCCG ---((((((((((---((.....))....((..(((.((((...((..((((((((((........))))))))))..))..)))))))...)).......)))))..-.....))))). ( -51.11) >DroPer_CAF1 10033 102 + 1 UCCGGCAGUCGGCAAUGGGAUCUCCACCAGCCAGCGGCCCUAUGUGGCGCCGAGGCUGCGCAACAAUGGCCUUGGCAUCAGUCGGGGAC---------------CGGUGUCACCUUU--- ....(((((((((..(((........))))))..((((((.....)).)))).))))))...(((.((((((((((....))))))).)---------------)).))).......--- ( -41.80) >consensus ___GGGAACGGGC___GGUAUCUCCACCAGCCAGCGUCCCUAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAU_UCAGAUUCCCG ...(((((..(((...((........)).)))..((.((((...((..((((((((((........))))))))))..))..)))).((((........)))).))........))))). (-35.21 = -36.68 + 1.47)


| Location | 1,550,861 – 1,550,978 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -51.60 |
| Consensus MFE | -48.55 |
| Energy contribution | -48.92 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1550861 117 + 23771897 UAUGUGGCGCCCAGGCUGCGAAACAACGGCCUAGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAUUCAGAUUCCCGGGCAACCUGGCCUCGUGGGUAACAACAAUCCACAGCAG ..((.(((((((((((((........))))))..((....)).)))))))))((...(((((((((...........)))))...))))....((((((.......)))))).)).. ( -49.00) >DroSec_CAF1 8537 114 + 1 UAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAUUCAGAUUCCCGGGCAACCUGGCCUCGUGGGUAA---CAAUCCACAGCAG ..((((((((((((((((........)))))))))).....((((((.....)).......(((((...........)))))..)))))))..((((((..---..)))))).))). ( -52.30) >DroSim_CAF1 8655 114 + 1 UAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAUUCAGAUUCCCGGGCAACCUGGCCUCGUGGGUAA---CAAUCCACAGCAG ..((((((((((((((((........)))))))))).....((((((.....)).......(((((...........)))))..)))))))..((((((..---..)))))).))). ( -52.30) >DroYak_CAF1 9220 114 + 1 UAUGUGGCGCCCAGGCUGCGGAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGUCCGAUUCAGAUUCCCGGGCAACCUGGCCUCGUGGGAAA---CAACCCCCCACAG ...(.(((((((((((((........)))))))))).....((((..((((.(.....)(((..(......)...)))))))..))))))).)(((((...---......))))).. ( -52.80) >consensus UAUGUGGCGCCCAGGCUGCGAAACAACGGCCUGGGCAUCAGCAGGGCGCCCAGCACUCCGGGCCCGAUUCAGAUUCCCGGGCAACCUGGCCUCGUGGGUAA___CAAUCCACAGCAG ..((((((((((((((((........)))))))))).....((((..((((.(.....)(((..(......)...)))))))..)))))))..((((((.......)))))).))). (-48.55 = -48.92 + 0.38)

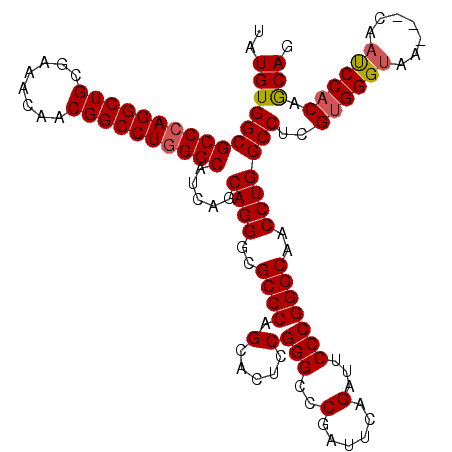
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:14 2006