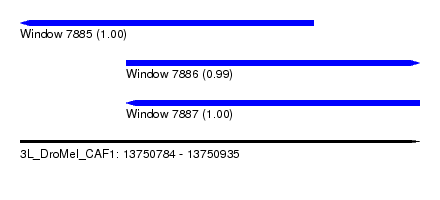
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,750,784 – 13,750,935 |
| Length | 151 |
| Max. P | 0.999720 |

| Location | 13,750,784 – 13,750,895 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -30.39 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
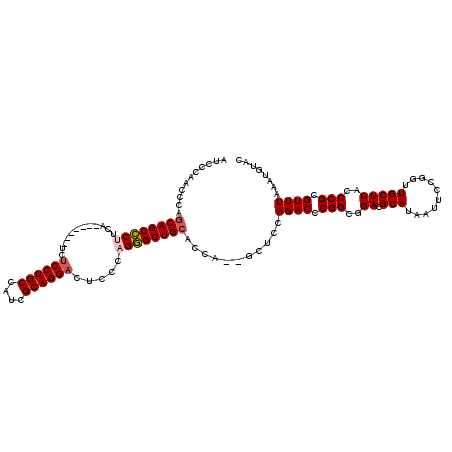
>3L_DroMel_CAF1 13750784 111 - 23771897 AUCCCAACCCAGCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ...........((((((((...-------..((((((....)))))).....))))))))....--.....((((.(((..((.(((..........)))))..))).))))........ ( -37.50) >DroSec_CAF1 157526 111 - 1 AUCCCAACCCACCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ............(((((((...-------..((((((....)))))).....))))))).....--.....((((.(((..((.(((..........)))))..))).))))........ ( -32.90) >DroSim_CAF1 169768 111 - 1 AUCCCAACCCAGCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ...........((((((((...-------..((((((....)))))).....))))))))....--.....((((.(((..((.(((..........)))))..))).))))........ ( -37.50) >DroEre_CAF1 164382 117 - 1 AUCCCAACCCUGCAGCCCCACAUCGCAGG-ACCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUUCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ..........((((((((......((((.-...)))).......((....)).))))))))...--.....((((.(((..((.(((..........)))))..))).))))........ ( -34.50) >DroYak_CAF1 168711 120 - 1 AUCCCAACCCAGCAGCUCCUCCUCCUUCAUCUCCUGCCGUCGCAGGACUCCCAGAGCUGCACCAGAGCUCCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ...........(((((((.............((((((....))))))......)))))))....((((.((.((....((....)).....)).))..))))....((((....)))).. ( -30.81) >consensus AUCCCAACCCAGCAGCCCUUCA_______UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA__GCUCCAGACCCGGCGGACGCAUAAUUCCGGUUGCUCACCCGCGUUUAAAUGUAC ...........((((((((............((((((....)))))).....))))))))...........((((.(((..((.(((..........)))))..))).))))........ (-30.39 = -30.87 + 0.48)

| Location | 13,750,824 – 13,750,935 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -45.67 |
| Consensus MFE | -37.89 |
| Energy contribution | -37.57 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
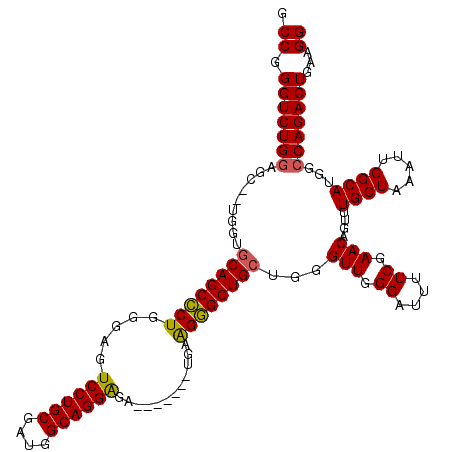
>3L_DroMel_CAF1 13750824 111 + 23771897 GCCGGGUCUGGAGC--UGGUGCAGCCCUGGGAGUCCUGCGAUGGCAGGAGA-------UGAAGGGCUGCUGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.(((((((.((--..((((((((((.....((((((....))))))..-------...))))))))...(((.((....)).)))..........))..))....)))))))...)) ( -46.80) >DroSec_CAF1 157566 111 + 1 GCCGGGUCUGGAGC--UGGUGCAGCCCUGGGAGUCCUGCGAUGGCAGGAGA-------UGAAGGGCUGGUGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.(((((((.((--..((.(((((((.....((((((....))))))..-------...)))))))(..(..((...........))..)..)...))..))....)))))))...)) ( -44.80) >DroSim_CAF1 169808 111 + 1 GCCGGGUCUGGAGC--UGGUGCAGCCCUGGGAGUCCUGCGAUGGCAGGAGA-------UGAAGGGCUGCUGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.(((((((.((--..((((((((((.....((((((....))))))..-------...))))))))...(((.((....)).)))..........))..))....)))))))...)) ( -46.80) >DroEre_CAF1 164422 117 + 1 GCCGGGUCUGAAGC--UGGUGCAGCCCUGGGAGUCCUGCGAUGGCAGGGU-CCUGCGAUGUGGGGCUGCAGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.((((((((((--((.(.((((((((.(.((((..(.((.((((...-.)))).)))..))))).)))))))).).........))))))(((.....))).....))))))...)) ( -49.10) >DroYak_CAF1 168751 120 + 1 GCCGGGUCUGGAGCUCUGGUGCAGCUCUGGGAGUCCUGCGACGGCAGGAGAUGAAGGAGGAGGAGCUGCUGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.((((((((((((....((((((((.....((((((....))))))............)))))))).))))).................((((.....))))...)))))))...)) ( -40.83) >consensus GCCGGGUCUGGAGC__UGGUGCAGCCCUGGGAGUCCUGCGAUGGCAGGAGA_______UGAAGGGCUGCUGGGUUGGGAUUUUCGAACAGUUUGCUAAAUUGGCAUGGCCAGACUGAAGG .((.(((((((.........((((((((.....((((((....))))))............))))))))...(((.((....)).)))....((((.....))))...)))))))...)) (-37.89 = -37.57 + -0.32)


| Location | 13,750,824 – 13,750,935 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -29.13 |
| Energy contribution | -29.45 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13750824 111 - 23771897 CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCAGCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGC ......((((((...(((.......))).......................((((((((...-------..((((((....)))))).....))))))))....--...))))))..... ( -36.50) >DroSec_CAF1 157566 111 - 1 CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCACCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGC ......((((((...(((.......)))........................(((((((...-------..((((((....)))))).....))))))).....--...))))))..... ( -31.90) >DroSim_CAF1 169808 111 - 1 CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCAGCAGCCCUUCA-------UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUCCAGACCCGGC ......((((((...(((.......))).......................((((((((...-------..((((((....)))))).....))))))))....--...))))))..... ( -36.50) >DroEre_CAF1 164422 117 - 1 CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCUGCAGCCCCACAUCGCAGG-ACCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA--GCUUCAGACCCGGC ......((((((...(((.......)))......................((((((((......((((.-...)))).......((....)).))))))))...--...))))))..... ( -31.40) >DroYak_CAF1 168751 120 - 1 CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCAGCAGCUCCUCCUCCUUCAUCUCCUGCCGUCGCAGGACUCCCAGAGCUGCACCAGAGCUCCAGACCCGGC ......((((((...(((.......)))....(((((.....)........(((((((.............((((((....))))))......)))))))....)))).))))))..... ( -30.91) >consensus CCUUCAGUCUGGCCAUGCCAAUUUAGCAAACUGUUCGAAAAUCCCAACCCAGCAGCCCUUCA_______UCUCCUGCCAUCGCAGGACUCCCAGGGCUGCACCA__GCUCCAGACCCGGC ......((((((...(((.......))).......................((((((((............((((((....)))))).....)))))))).........))))))..... (-29.13 = -29.45 + 0.32)

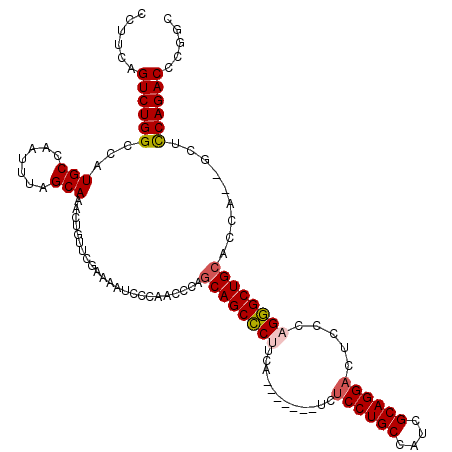
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:55 2006