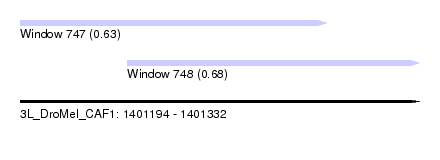
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,401,194 – 1,401,332 |
| Length | 138 |
| Max. P | 0.683233 |

| Location | 1,401,194 – 1,401,300 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -23.13 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1401194 106 + 23771897 CCAGCUGGAGUACAGGCGCUAUUUCUGGUGGCACUCCAUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGUCUCCUGGG ((((..((((((..(((((.......(((((....)))))....(((((((........))))))).....)))))...))))))((........))....)))). ( -37.60) >DroGri_CAF1 31676 100 + 1 GCCGCAGCUGUACGAGCAAUUCUUCUGGUGGCAUUCCAUGUAUCGACUCCACAAGAGUCCGCAUCCCUUGUGCGCCCUCUGCCGCCAACUCGGU------CGCGAG ..(((.((((...(((......)))((((((((...........(((((.....)))))(((((.....))))).....))))))))...))))------.))).. ( -31.60) >DroSec_CAF1 56099 106 + 1 CCCGCUGGAGUACAGGCGCUACUUCUGGUGGCACUCCAUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGG .((.(((((((...(((((.......(((((....)))))....(((((((........))))))).....)))))..(((.............)))))))))))) ( -40.62) >DroSim_CAF1 52229 106 + 1 UCCGCUGGAGUACAGGCGCUACUUCUGGUGGCACUCCAUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGG .((.(((((((...(((((.......(((((....)))))....(((((((........))))))).....)))))..(((.............)))))))))))) ( -40.42) >DroEre_CAF1 57766 106 + 1 CCCGGAGGAGUACAGGCGCUACUUCUGGUGGCAUGCCAUCUACAAGCUGCGCCAGGCCUCGCAACCCUACUGCGCCCUCUGCUCCCUGGUGCAACAGGCUCCAGGG ..((((((.((...(((((..(((..(((((....)))))...)))..))))).......(((.......)))))))))))..((((((.((.....)).)))))) ( -45.60) >DroYak_CAF1 59504 106 + 1 UCCAAAGGAGUACAGGCGCUACUUCUGGUGGCACUCCAUCUACAAGCUGCGCCACGCCUCGCAACCCUACUGCGCCCUCUGCUCCCUCGUGCAAAAGGCUCCAGGG .((...(((.....(((((..(((..(((((....)))))...)))..)))))..((((((((.......)))).....(((........)))..)))))))..)) ( -32.60) >consensus CCCGCAGGAGUACAGGCGCUACUUCUGGUGGCACUCCAUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGG ......((((((.((((((..(((..(((((....)))))...)))..)))).......(((((.....)))))..)).))))))..................... (-23.13 = -23.88 + 0.75)

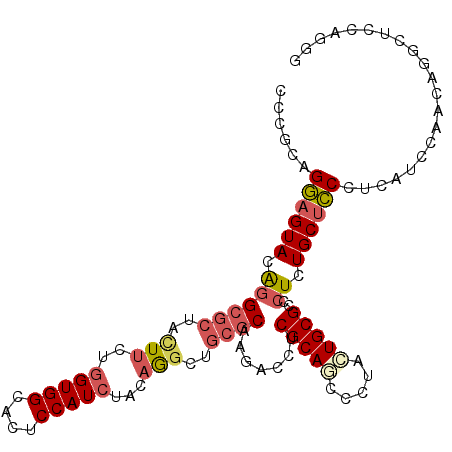
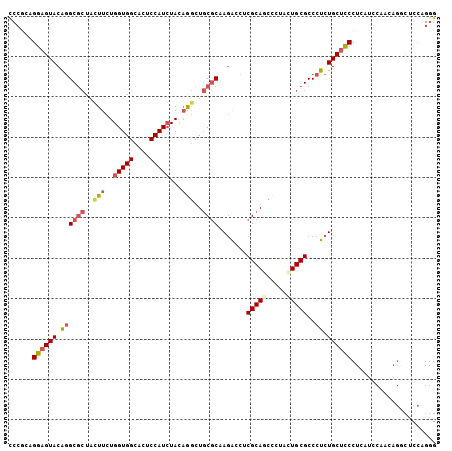
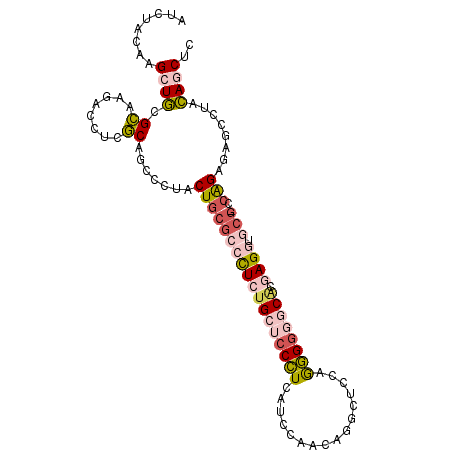
| Location | 1,401,231 – 1,401,332 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.64 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1401231 101 + 23771897 AUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGUCUCCUGGGGGGCACGAGGUGCG-CCAGAGAUCCUACAGCUU ((((...(((((((........)))))))...(((((((((((((((((.((.............))))))))).)))).)))-.))))))).......... ( -41.42) >DroSec_CAF1 56136 101 + 1 AUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGGGGGCACGAGGUGCG-CCAGAGAGCCUACAGCUC .......(((((((........)))))))...((((((((((((((((((...((....)).....)))))))).)))).)))-.))).((((.....)))) ( -45.90) >DroSim_CAF1 52266 101 + 1 AUCUACAGGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGGGGGCACGAGGUGCG-CCAGAGAGCCUACAGCUC .......(((((((........)))))))...((((((((((((((((((...((....)).....)))))))).)))).)))-.))).((((.....)))) ( -45.90) >DroEre_CAF1 57803 101 + 1 AUCUACAAGCUGCGCCAGGCCUCGCAACCCUACUGCGCCCUCUGCUCCCUGGUGCAACAGGCUCCAGGGGGGCACAAGAUCCG-CCAGAGAUCCGACAUCAC ........(.(((..((((...((((.......))))...))))((((((((.((.....)).))))))))))))..(((((.-...).))))......... ( -30.80) >DroYak_CAF1 59541 101 + 1 AUCUACAAGCUGCGCCACGCCUCGCAACCCUACUGCGCCCUCUGCUCCCUCGUGCAAAAGGCUCCAGGGGCACCCGAGAUCCG-UCAGAGAUCCUACAGCAC ........((((......((((((((.......)))(((...(((........)))...)))....)))))......(((((.-...).))))...)))).. ( -27.90) >DroAna_CAF1 59762 102 + 1 AUCUACAAGAUCCGGGCCAUCGCCCAGCCGUACUGCGCCUUGUGCCACUUCAUCCAAACGCCCCUGAAGGACCGUAAGGCGAAGAAGGAGAGCACGGACCUU ......(((.(((((((....)))).(((...((.(((((((((...(((((............)))))...)))))))))....))..).))..))).))) ( -30.50) >consensus AUCUACAAGCUGCGCAAGACCUCGCAGCCCUACUGCGCCCUCUGCUCCCUCAUCCAACAGGCUCCAGGGGGGCACGAGGUGCG_CCAGAGAGCCUACAGCUC ........((((.((........)).......((((((((((((((((((................)))))))).)))).)))..)))........)))).. (-18.16 = -19.64 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:35 2006