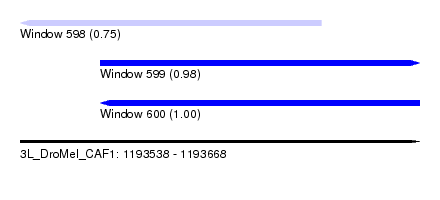
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,193,538 – 1,193,668 |
| Length | 130 |
| Max. P | 0.996731 |

| Location | 1,193,538 – 1,193,636 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.81 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -34.39 |
| Energy contribution | -34.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1193538 98 - 23771897 GCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAGGCUCAGGGCCCCUCCAUGCCAUUAUUGCCCCAGUGCGGUU (((....(((..((.---(((((((((....)))).(((((((((((.....)))))).)))))..)))))...))..)))......((......))))). ( -36.10) >DroSec_CAF1 9180 98 - 1 GCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAGGCUCAGGGCCCCUCCAUGCCAUUAUUGCCCCAGUGCGGUU (((....(((..((.---(((((((((....)))).(((((((((((.....)))))).)))))..)))))...))..)))......((......))))). ( -36.10) >DroSim_CAF1 8578 98 - 1 GCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAAGCUUAGGGGCCCUCCAUGCCAUUAUUGCCCCAGUGCGGUU (((....(((..((.---(((((...((((((.........((((((.....))))))))))))...)))))..))..)))......((......))))). ( -38.90) >DroYak_CAF1 8748 101 - 1 GCCCCUCGGCUCGGUGGUGGCCCAUUGGCUUCAAUUAGUCUGGGCCCAACAAAGGCCCGAAGCUCAGGGCCUCUCCAUGCCAUUAUUGCCCCAGUGCGGUU ((((((.(((..((((((((((((..((((......))))))))))......((((((........))))))......))))))...)))..)).).))). ( -39.50) >consensus GCCUCUCGGCUCGGU___GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAAGCUCAGGGCCCCUCCAUGCCAUUAUUGCCCCAGUGCGGUU (((....(((..((....(((((...((((((.........((((((.....))))))))))))..)))))...))..)))......((......))))). (-34.39 = -34.45 + 0.06)

| Location | 1,193,564 – 1,193,668 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -33.97 |
| Energy contribution | -33.60 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1193564 104 + 23771897 GAGGGGCCCUGAGCCUCGGGCCUUUGUUAGACCCAGACUAAUUGAAGCCAAUGGGCC---ACCGAGCCGAGAGGCUGAAGGGCCAUAAUUGAGAUCUGAGAUCAAAA ....((((((.((((((.(((.((.(((((.......))))).)).)))....(((.---.....)))..))))))..))))))........((((...)))).... ( -39.30) >DroSec_CAF1 9206 104 + 1 GAGGGGCCCUGAGCCUCGGGCCUUUGUUAGACCCAGACUAAUUGAAGCCAAUGGGCC---ACCGAGCCGAGAGGCUGAAGGGCCAUAAUUGAGAUCUGAGAUCAAAA ....((((((.((((((.(((.((.(((((.......))))).)).)))....(((.---.....)))..))))))..))))))........((((...)))).... ( -39.30) >DroSim_CAF1 8604 104 + 1 GAGGGCCCCUAAGCUUCGGGCCUUUGUUAGACCCAGACUAAUUGAAGCCAAUGGGCC---ACCGAGCCGAGAGGCUGAAGGGCCAUAAUUGAGAUCUGAGAUCAAAA ((((((((.........))))))))....((.(((((((.((((....)))).((((---....((((....))))....)))).......)).)))).).)).... ( -35.50) >DroYak_CAF1 8774 100 + 1 GAGAGGCCCUGAGCUUCGGGCCUUUGUUGGGCCCAGACUAAUUGAAGCCAAUGGGCCACCACCGAGCCGAGGGGCUGAAGGGCCAUAAUUGA-------GAUCAAAA ....((((((.((((((.(((.((.((.(((((((..((......))....)))))).).)).)))))..))))))..))))))....(((.-------...))).. ( -39.70) >consensus GAGGGGCCCUGAGCCUCGGGCCUUUGUUAGACCCAGACUAAUUGAAGCCAAUGGGCC___ACCGAGCCGAGAGGCUGAAGGGCCAUAAUUGAGAUCUGAGAUCAAAA ....((((((.((((((.(((.((.(((((.......))))).)).)))....(((.........)))..))))))..))))))....((((.........)))).. (-33.97 = -33.60 + -0.37)

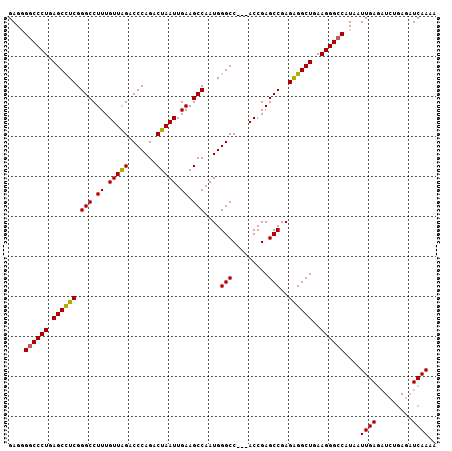
| Location | 1,193,564 – 1,193,668 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -37.04 |
| Energy contribution | -37.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1193564 104 - 23771897 UUUUGAUCUCAGAUCUCAAUUAUGGCCCUUCAGCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAGGCUCAGGGCCCCUC ....((((...))))........((((((..((((((..((((..((---((((((..((((......))))))))))..))...)))).)))))).)))))).... ( -42.30) >DroSec_CAF1 9206 104 - 1 UUUUGAUCUCAGAUCUCAAUUAUGGCCCUUCAGCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAGGCUCAGGGCCCCUC ....((((...))))........((((((..((((((..((((..((---((((((..((((......))))))))))..))...)))).)))))).)))))).... ( -42.30) >DroSim_CAF1 8604 104 - 1 UUUUGAUCUCAGAUCUCAAUUAUGGCCCUUCAGCCUCUCGGCUCGGU---GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAAGCUUAGGGGCCCUC ....((((...))))........(((((((.(((.((..((((..((---((((((..((((......))))))))))..))...)))).)).))).)))))))... ( -38.10) >DroYak_CAF1 8774 100 - 1 UUUUGAUC-------UCAAUUAUGGCCCUUCAGCCCCUCGGCUCGGUGGUGGCCCAUUGGCUUCAAUUAGUCUGGGCCCAACAAAGGCCCGAAGCUCAGGGCCUCUC ........-------........((((((..(((...((((..(..((..((((((..((((......))))))))))...))..)..)))).))).)))))).... ( -38.60) >consensus UUUUGAUCUCAGAUCUCAAUUAUGGCCCUUCAGCCUCUCGGCUCGGU___GGCCCAUUGGCUUCAAUUAGUCUGGGUCUAACAAAGGCCCGAAGCUCAGGGCCCCUC .......................((((((..((((((..((((..((...((((((..((((......))))))))))..))...)))).)))))).)))))).... (-37.04 = -37.85 + 0.81)


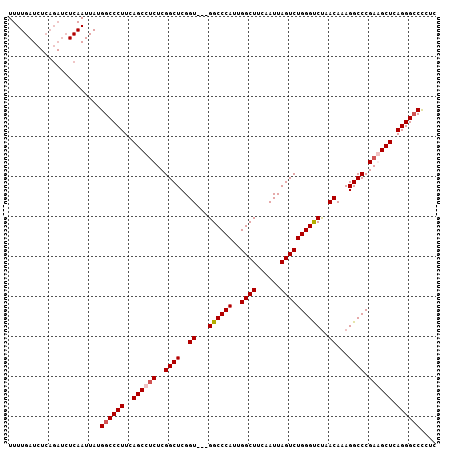
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:10 2006