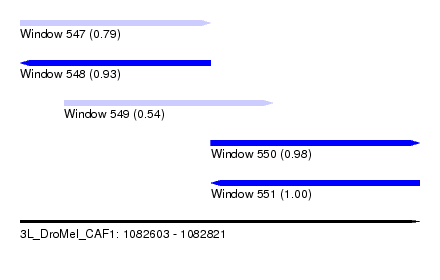
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,082,603 – 1,082,821 |
| Length | 218 |
| Max. P | 0.997046 |

| Location | 1,082,603 – 1,082,707 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.34 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1082603 104 + 23771897 ------UGAAAUUUAAG---UUG------C-UUAGAUUCACUCACCCUUUCUUCCUCUCGGAUCAUCUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCC ------((((.((((((---...------)-)))))))))...................(((((..((((.(((....(((((((.....)))))))))).))))....)))))...... ( -27.90) >DroSim_CAF1 58465 104 + 1 ------UGCAAUUUAAG---UGG------C-UUUGAUUCACUCACCCUUUCUUCCUCUCGGAUCAUCUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCC ------..........(---(((------.-..(((.....)))...............(((((..((((.(((....(((((((.....)))))))))).))))....))))).)))). ( -28.40) >DroEre_CAF1 63998 101 + 1 ------UC---UCUUAG---GGU------C-UUGGAUUCACUCACCCUUUCUUCCUCUCGGAUCAUUUCGGCGAUCCCAGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCGCCGCC ------..---......---(((------.-.(((((((.(((.((.............(((((........)))))..((((((.....))))))...)).)))...)))))))..))) ( -29.40) >DroWil_CAF1 90539 96 + 1 ------U--------------GAUUUUCUG-UUGG---AACUUACCCGCUCUUCUUCACGUAUCAUUUCAGCUAUGCCGGGCUUGAUCUCCAUAUCAUCGGCUAAUGAAGACGACGCUGU ------(--------------((.((((..-..))---)).)))...((((.((((((.(((.(......).)))(((((...((((......)))))))))...)))))).)).))... ( -16.80) >DroYak_CAF1 59400 93 + 1 ------U--------------GC------U-CUAGAUUCACUCACCCUUUCUUCCUCUCGGAUCAUUUCCGCGAUCCCUGGCUUGAUCUCCAUGUCAUCGGCGAGCGAGGAUCCCCCGCC ------.--------------((------.-...((.....)).........(((((..(((((........)))))((.(((((((......))))..))).)).)))))......)). ( -19.70) >DroAna_CAF1 58324 114 + 1 UUGUUUUUAAAUUUAAAAAAUUG------UAAAACUUUGACUUACCCUUUCUUCCUCUCGAAUCAUUUCGGCGAUUCCCGGCUUGAUCUCCAGGUCAUCGGCUAGAGAGGAUCCUCCUCC ..(((((..((((.....)))).------.))))).................((((((((((((........)))))..(((((((((....)))))..)))).)))))))......... ( -24.90) >consensus ______U____UUUAAG___UGG______C_UUAGAUUCACUCACCCUUUCUUCCUCUCGGAUCAUUUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCC .................................................((((((((.((((((........)))))..((((((.....))))))....).))).)))))......... (-17.45 = -17.45 + 0.00)

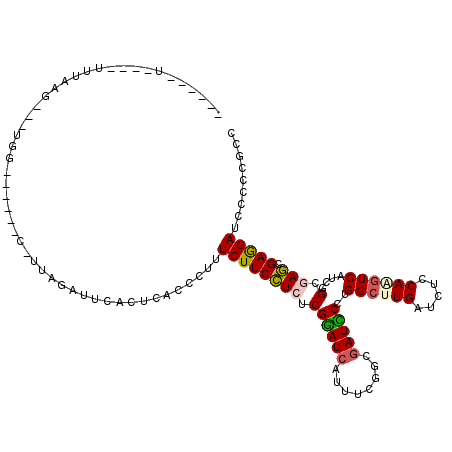
| Location | 1,082,603 – 1,082,707 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -18.06 |
| Energy contribution | -19.57 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1082603 104 - 23771897 GGCGGGGGAUCCUCGCUCGCCGAUGACUUGGAGAUCAAGCCAGGGAUCGCCGAGAUGAUCCGAGAGGAAGAAAGGGUGAGUGAAUCUAA-G------CAA---CUUAAAUUUCA------ .((...((((..(((((((((.....(((((.((((........)))).)))))....(((....)))......)))))))))))))..-)------)..---...........------ ( -37.60) >DroSim_CAF1 58465 104 - 1 GGCGGGGGAUCCUCGCUCGCCGAUGACUUGGAGAUCAAGCCAGGGAUCGCCGAGAUGAUCCGAGAGGAAGAAAGGGUGAGUGAAUCAAA-G------CCA---CUUAAAUUGCA------ (((....(((..(((((((((.....(((((.((((........)))).)))))....(((....)))......))))))))))))...-)------)).---...........------ ( -39.10) >DroEre_CAF1 63998 101 - 1 GGCGGCGGAUCCUCGCUCGCCGAUGACUUGGAGAUCAAGCCUGGGAUCGCCGAAAUGAUCCGAGAGGAAGAAAGGGUGAGUGAAUCCAA-G------ACC---CUAAGA---GA------ ((((((((....)))).)))).....((((((..(((.((((((.....)).......(((....))).....))))...))).)))))-)------...---......---..------ ( -34.50) >DroWil_CAF1 90539 96 - 1 ACAGCGUCGUCUUCAUUAGCCGAUGAUAUGGAGAUCAAGCCCGGCAUAGCUGAAAUGAUACGUGAAGAAGAGCGGGUAAGUU---CCAA-CAGAAAAUC--------------A------ ...((.((.(((((((..((((.((((......))))....))))....(......)....))))))).))))(((.....)---))..-.........--------------.------ ( -19.50) >DroYak_CAF1 59400 93 - 1 GGCGGGGGAUCCUCGCUCGCCGAUGACAUGGAGAUCAAGCCAGGGAUCGCGGAAAUGAUCCGAGAGGAAGAAAGGGUGAGUGAAUCUAG-A------GC--------------A------ .((...((((..((((((((((((..(.(((........))))..)))..........(((....)))......)))))))))))))..-.------))--------------.------ ( -31.80) >DroAna_CAF1 58324 114 - 1 GGAGGAGGAUCCUCUCUAGCCGAUGACCUGGAGAUCAAGCCGGGAAUCGCCGAAAUGAUUCGAGAGGAAGAAAGGGUAAGUCAAAGUUUUA------CAAUUUUUUAAAUUUAAAAACAA .........(((((((..(.((((..(((((........))))).)))).)(((....))))))))))(((((..(((((.......))))------)..)))))............... ( -27.60) >consensus GGCGGGGGAUCCUCGCUCGCCGAUGACUUGGAGAUCAAGCCAGGGAUCGCCGAAAUGAUCCGAGAGGAAGAAAGGGUGAGUGAAUCUAA_G______CCA___CUUAAA____A______ ............((((((((((((..(((((........))))).)))..........(((....)))......)))))))))..................................... (-18.06 = -19.57 + 1.50)

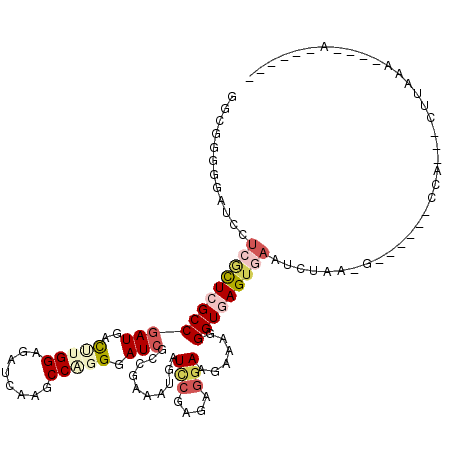

| Location | 1,082,627 – 1,082,741 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1082627 114 + 23771897 CUCACCCUUUCUUCCUCUCGGAUCAUCUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCCUGAACCCACUCCGCCUGCACCUC------CUCCUCCAACU ...................(((....((((.(((....(((((((.....)))))))))).)))).(((((......((..((......)).)).......))------))).))).... ( -26.12) >DroSim_CAF1 58489 114 + 1 CUCACCCUUUCUUCCUCUCGGAUCAUCUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCCUGAACCCACUCCGCCUGCACCUC------CUCCUCCAACU ...................(((....((((.(((....(((((((.....)))))))))).)))).(((((......((..((......)).)).......))------))).))).... ( -26.12) >DroEre_CAF1 64019 120 + 1 CUCACCCUUUCUUCCUCUCGGAUCAUUUCGGCGAUCCCAGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCGCCGCCCGAACCCACUCCGCCUGCAGCUCCUCCUCCUCCCCCCACU ...................(((((........)))))..((((((.....))))))...((.(((.(((((...((.((.((.........))...)).))...))))))))))...... ( -29.70) >DroYak_CAF1 59413 114 + 1 CUCACCCUUUCUUCCUCUCGGAUCAUUUCCGCGAUCCCUGGCUUGAUCUCCAUGUCAUCGGCGAGCGAGGAUCCCCCGCCUGCACCUACUCCGCCUGCACCUG------CGCCGCCUACU ...............((.((((.....)))).)).....(((.((((......))))..((((.(((.((....))))).((((...........))))....------))))))).... ( -26.40) >consensus CUCACCCUUUCUUCCUCUCGGAUCAUCUCGGCGAUCCCUGGCUUGAUCUCCAAGUCAUCGGCGAGCGAGGAUCCCCCGCCUGAACCCACUCCGCCUGCACCUC______CUCCUCCAACU ...................(((((........)))))..((((((.....))))))...((((((.(.((.((........)).))).)).))))......................... (-21.64 = -21.95 + 0.31)

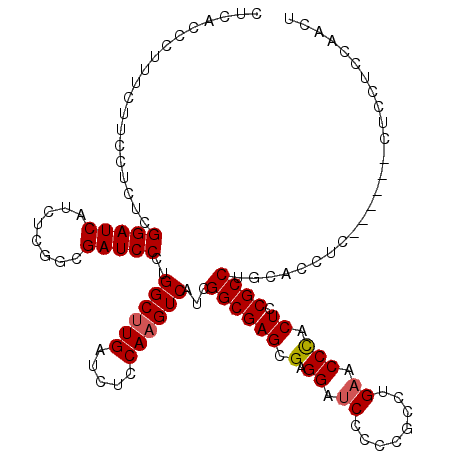
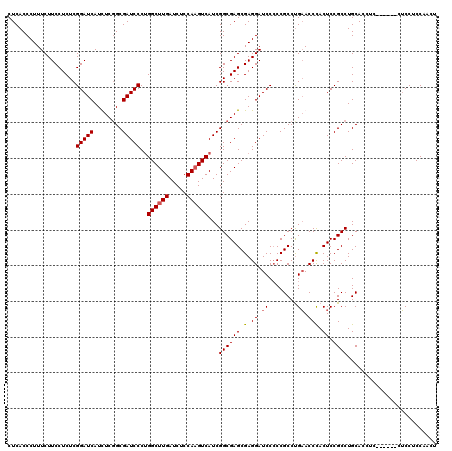
| Location | 1,082,707 – 1,082,821 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -58.97 |
| Consensus MFE | -44.30 |
| Energy contribution | -44.43 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1082707 114 + 23771897 UGAACCCACUCCGCCUGCACCUC------CUCCUCCAACUCCCCCGCCGCCCACUCCACCACCGCCAUGGUGGUGCGGCGGUGGAGGCGGAAGGCGUGGCUCGGCGGACGGACCUGGUCC .........((((((.((..(.(------((..(((..((((.(((((((.....((((((......)))))).))))))).))))..)))))).)..))..)))))).((((...)))) ( -56.00) >DroSim_CAF1 58569 114 + 1 UGAACCCACUCCGCCUGCACCUC------CUCCUCCAACUCCCCCACCGCCAACUCCACCACCGCCAUGGUGGUGCGGCGGUGGAGGCGGUAGGCGUGGCUCGGUGGACGGACCAGGUCC .....(((((..(((.((.((((------(.(((((.........((((((.....(((((((.....))))))).))))))))))).)).))).)))))..)))))..((((...)))) ( -53.10) >DroEre_CAF1 64099 120 + 1 CGAACCCACUCCGCCUGCAGCUCCUCCUCCUCCCCCCACUCCACCGCCACCCACUCCACCACCGCCAUGGUGGUGCGGCGGCGGAGGCGGAAGGCGUGGCUCAGUGGGCGGACCAGGUCC ...(((...((((((..((((..(.(((..(((.....((((.(((((........(((((((.....))))))).))))).))))..)))))).)..)))..)..))))))...))).. ( -56.30) >DroYak_CAF1 59493 114 + 1 UGCACCUACUCCGCCUGCACCUG------CGCCGCCUACUCCCCCGCCACCCACUCCACCUCCGCCGUGGUGGUGCGGUGGUGGAGGAGGUAGGCGUGGCUCAGCGGGCGGACCGGGACC ....(((..(((((((((....(------(..(((((((..((((((((((((((((((.......)))).)))).)))))))).))..)))))))..))...)))))))))..)))... ( -70.50) >consensus UGAACCCACUCCGCCUGCACCUC______CUCCUCCAACUCCCCCGCCACCCACUCCACCACCGCCAUGGUGGUGCGGCGGUGGAGGCGGAAGGCGUGGCUCAGCGGACGGACCAGGUCC ....((...(((((((((....................(.((.((((((((.....(((((((.....))))))).)))))))).)).)...(((...)))..)))))))))...))... (-44.30 = -44.43 + 0.13)

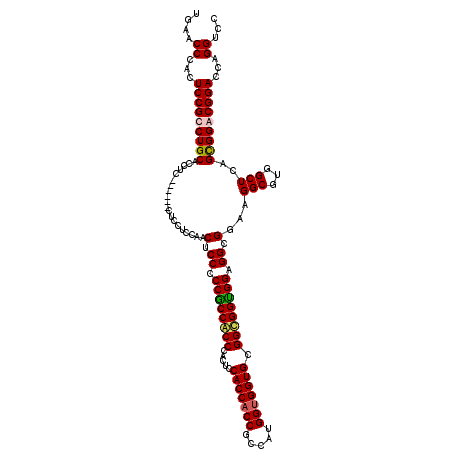
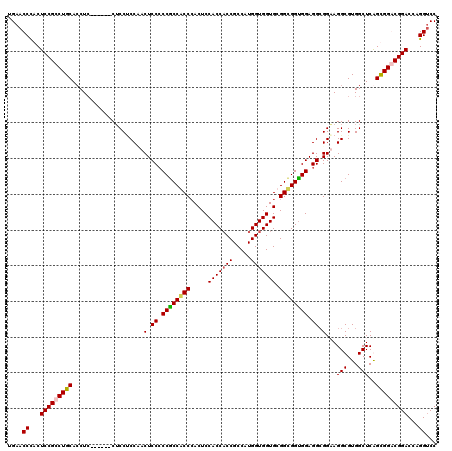
| Location | 1,082,707 – 1,082,821 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Mean single sequence MFE | -69.90 |
| Consensus MFE | -58.28 |
| Energy contribution | -59.46 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1082707 114 - 23771897 GGACCAGGUCCGUCCGCCGAGCCACGCCUUCCGCCUCCACCGCCGCACCACCAUGGCGGUGGUGGAGUGGGCGGCGGGGGAGUUGGAGGAG------GAGGUGCAGGCGGAGUGGGUUCA (((((.(.((((((((((...((...(((.(((.((((.(((((((.(((((((....))))))).....))))))).)))).)))))).)------).))))..)))))).).))))). ( -66.40) >DroSim_CAF1 58569 114 - 1 GGACCUGGUCCGUCCACCGAGCCACGCCUACCGCCUCCACCGCCGCACCACCAUGGCGGUGGUGGAGUUGGCGGUGGGGGAGUUGGAGGAG------GAGGUGCAGGCGGAGUGGGUUCA (((((((.((((((((((...((...(((.((.(((((((((((((.(((((((....))))))).).))))))))))))....))))).)------).))))..)))))).))))))). ( -71.10) >DroEre_CAF1 64099 120 - 1 GGACCUGGUCCGCCCACUGAGCCACGCCUUCCGCCUCCGCCGCCGCACCACCAUGGCGGUGGUGGAGUGGGUGGCGGUGGAGUGGGGGGAGGAGGAGGAGCUGCAGGCGGAGUGGGUUCG (((((((.((((((..(.(..((.(.((((((.(((((((((((((.(((((((....))))))).....)))))))))))..)).)))))).)..))..).)..)))))).))))))). ( -74.80) >DroYak_CAF1 59493 114 - 1 GGUCCCGGUCCGCCCGCUGAGCCACGCCUACCUCCUCCACCACCGCACCACCACGGCGGAGGUGGAGUGGGUGGCGGGGGAGUAGGCGGCG------CAGGUGCAGGCGGAGUAGGUGCA .(..((..((((((((((..((..(((((((.((((((.(((((((.(((((........))))).)).))))).)))))))))))))..)------).))))..))))))...))..). ( -67.30) >consensus GGACCUGGUCCGCCCACCGAGCCACGCCUACCGCCUCCACCGCCGCACCACCAUGGCGGUGGUGGAGUGGGCGGCGGGGGAGUUGGAGGAG______GAGGUGCAGGCGGAGUGGGUUCA (((((((.((((((((((........((((((((((((((((((((.(((((((....))))))).)).)))))))))))...)))))).)........))))..)))))).))))))). (-58.28 = -59.46 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:22 2006