| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
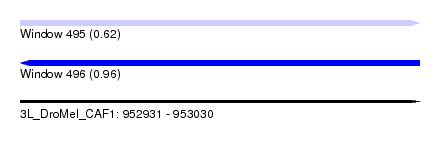
| Location | 952,931 – 953,030 |
| Length | 99 |
| Max. P | 0.960286 |

| Location | 952,931 – 953,030 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.51 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -17.84 |
| Energy contribution | -21.03 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
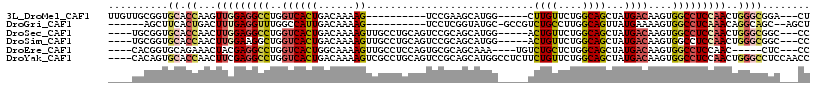
>3L_DroMel_CAF1 952931 99 + 23771897 AG---UCCGCCCAGUUGGAGGCCACUUGUCAUAGCUGCCAGAACAAG-----CCAUGCUUCGGA----------CUUUUGUCAGUGACCAGGCCUCCAACUUGGUGCACCGCAACAA .(---(.((((.(((((((((((....((((((((.((........)-----)...)))...((----------(....))).)))))..))))))))))).)))).))........ ( -39.10) >DroGri_CAF1 98329 98 + 1 AGCU--GCUGCCUGUUUGAGGCCACUUUUCAUAACUGCCAAGGCAGACGGC-GCAUACCGAGGA----------CUUUUGUCAAUGGCCAAACCUCAAAGUCAGUGAAGCU------ ((((--((((.((..((((((.............((((....))))..(((-.(((......((----------(....))).))))))...)))))))).))))..))))------ ( -29.60) >DroSec_CAF1 88349 105 + 1 GG---GCCGCCCAGUUGGAGGCCACUUGUCAUAGCUGCCAGAACAGU-----CCAUGCUGCGGACUGCAGGCAACUUUUGUCAGUGACCAGGCCUCCAAGUUGGUGCACCGCA---- ((---..((((.(.(((((((((....(((((((.((((....((((-----((.......))))))..)))).)).......)))))..))))))))).).))))..))...---- ( -46.11) >DroSim_CAF1 79796 105 + 1 GG---GCCGCCCAGUUGGAGGCCACUUGUCAUAGCUGCCAGAACAGU-----CCAUGCUGCGGACUGCAGGCAACUUUUGUCAGUGACCAGCCUUCCAAGUUGGUGCACCGCA---- ((---..((((.(.((((((((((((.(.((.((.((((....((((-----((.......))))))..)))).))..)).)))))....))).))))).).))))..))...---- ( -40.70) >DroEre_CAF1 89774 101 + 1 GG---GAG-----GUUGGAGGCCACUUGUCAUAGCUGCCAGAGCAGACA----UUUGCUGCGCACUGGAGGCAACUUUUGCCAGUGACCAGGCCUCGUAGUUUCUGCACCGUG---- ..---..(-----((..((((((....(((((.....((((.((((.(.----...)))))...)))).((((.....)))).)))))..))))))((((...)))))))...---- ( -37.80) >DroYak_CAF1 92157 113 + 1 GGUUGGAGGCCCAGUUGGAGGCCACUUGUCAUAGCUGCCAGAACAGAAGAGGCCAUGCUGCGGACUGCAGGCGACUUUUGUCAGUGACCAGGCCUCGAAGUUGGUGCACUGUG---- ....(.((((((((((.((((((....(((...((((.....(((((((..(((.(((........))))))..))))))))))))))..)))))).)).)))).)).)).).---- ( -40.40) >consensus GG___GCCGCCCAGUUGGAGGCCACUUGUCAUAGCUGCCAGAACAGA_____CCAUGCUGCGGACUGCAGGCAACUUUUGUCAGUGACCAGGCCUCCAAGUUGGUGCACCGCA____ ........(((((((((((((((....(((((..(((......)))................(((...((....))...))).)))))..))))))))).)))).)).......... (-17.84 = -21.03 + 3.20)

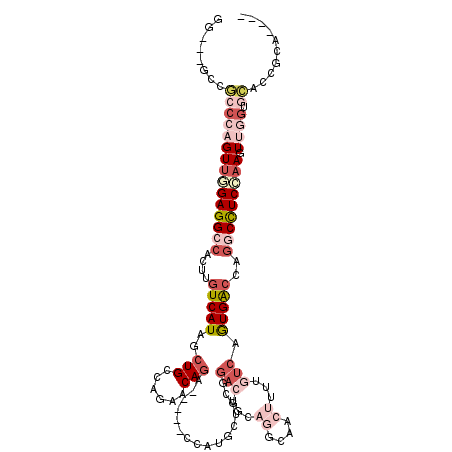
| Location | 952,931 – 953,030 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.51 |
| Mean single sequence MFE | -40.05 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 952931 99 - 23771897 UUGUUGCGGUGCACCAAGUUGGAGGCCUGGUCACUGACAAAAG----------UCCGAAGCAUGG-----CUUGUUCUGGCAGCUAUGACAAGUGGCCUCCAACUGGGCGGA---CU .......(((.(.((.(((((((((((..(((((.(((....)----------)).).(((...(-----((......))).))).))))....))))))))))).)).).)---)) ( -41.00) >DroGri_CAF1 98329 98 - 1 ------AGCUUCACUGACUUUGAGGUUUGGCCAUUGACAAAAG----------UCCUCGGUAUGC-GCCGUCUGCCUUGGCAGUUAUGAAAAGUGGCCUCAAACAGGCAGC--AGCU ------(((((((.((((((..((((.(((((((.(((....)----------))......))).-))))...))))..).)))))))))..((.((((.....)))).))--.))) ( -30.30) >DroSec_CAF1 88349 105 - 1 ----UGCGGUGCACCAACUUGGAGGCCUGGUCACUGACAAAAGUUGCCUGCAGUCCGCAGCAUGG-----ACUGUUCUGGCAGCUAUGACAAGUGGCCUCCAACUGGGCGGC---CC ----...(((((.(((..(((((((((..((((........(((((((.((((((((.....)))-----)))))...))))))).))))....))))))))).))))).))---). ( -51.10) >DroSim_CAF1 79796 105 - 1 ----UGCGGUGCACCAACUUGGAAGGCUGGUCACUGACAAAAGUUGCCUGCAGUCCGCAGCAUGG-----ACUGUUCUGGCAGCUAUGACAAGUGGCCUCCAACUGGGCGGC---CC ----...(((((.(((..(((((.....(((((((......(((((((.((((((((.....)))-----)))))...)))))))......)))))))))))).))))).))---). ( -46.50) >DroEre_CAF1 89774 101 - 1 ----CACGGUGCAGAAACUACGAGGCCUGGUCACUGGCAAAAGUUGCCUCCAGUGCGCAGCAAA----UGUCUGCUCUGGCAGCUAUGACAAGUGGCCUCCAAC-----CUC---CC ----...(((......)))..((((((..((((.((((....((((((......).)))))...----((((......))))))))))))....))))))....-----...---.. ( -32.90) >DroYak_CAF1 92157 113 - 1 ----CACAGUGCACCAACUUCGAGGCCUGGUCACUGACAAAAGUCGCCUGCAGUCCGCAGCAUGGCCUCUUCUGUUCUGGCAGCUAUGACAAGUGGCCUCCAACUGGGCCUCCAACC ----...(((......)))..((((((..((....((((.(((..((((((........))).)))..))).)))).(((..(((((.....)))))..)))))..))))))..... ( -38.50) >consensus ____UGCGGUGCACCAACUUGGAGGCCUGGUCACUGACAAAAGUUGCCUGCAGUCCGCAGCAUGG_____UCUGUUCUGGCAGCUAUGACAAGUGGCCUCCAACUGGGCGGC___CC ..........((.((...(((((((((..((((((......))............................((((....))))...))))....)))))))))..))))........ (-17.75 = -18.50 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:26 2006