| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 509,318 – 509,430 |
| Length | 112 |
| Max. P | 0.910884 |

| Location | 509,318 – 509,430 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
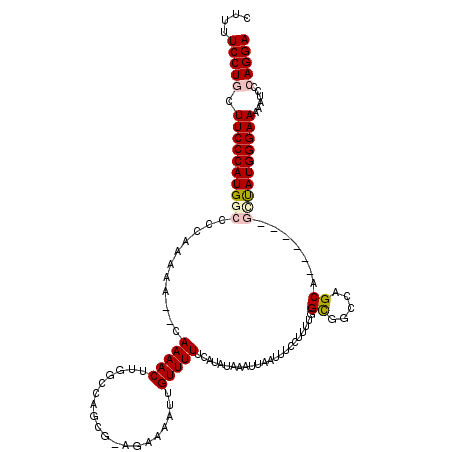
>3L_DroMel_CAF1 509318 112 - 23771897 CUUUUCCUGCUUCCCAUGGCCCCAAAAA--CAAAACUUGGCCAGCGUAGAAAAUUGUUUUUCAUAUAAAUUAAUUUCCUUUUGGCGGCCAGCA------GUUAUGGGAAAAAUCCCAGGA ....(((((.((((((((((........--......((((((.((..((.((((((.(((......))).)))))).))....))))))))..------))))))))))......))))) ( -27.59) >DroPse_CAF1 171212 104 - 1 UUUUUCCUGCUUCCCAUGGCCGAAAAA----AAAACC--CCCAAC--------AUGUUUU--CUAUAAAUUAAUUUCCUUUUUGUGAAAAGCAUGAAAAAGCAUGGGAAAAGUCCAAGGA ....((((((((((((((...(.....----......--..)..(--------(((((((--.((((((...........)))))).))))))))......)))))))..)))...)))) ( -21.82) >DroSec_CAF1 89494 112 - 1 CUUUUCCUGCUUCCCAUGGCCCCAAAAA--CAAAACUUGGCCAGCGAAGAAAAUUGUUUUUCAUAUAAAUUAAUUUCCUUUUGGCGGCCAGCA------GCUAUGGGAAAAAUGCCAGGA ....((((((((((((((((........--......((((((.((.((((((((((.(((......))).))))))..)))).))))))))..------))))))))))....).))))) ( -32.99) >DroEre_CAF1 92710 112 - 1 CUUUUCCUGCUUCCCAUGGCCCCAAAAA--CAAAACUUGGCCAGCGUAGAAAAUUGUUUUUCAUAUAAAUUAAUUUCCUUUUGGCGGCCAGCG------GCUAUGGGAAAAAUCCCAGGA ....(((((.(((((((((((.......--......((((((.((..((.((((((.(((......))).)))))).))....)))))))).)------))))))))))......))))) ( -33.54) >DroYak_CAF1 94823 114 - 1 CUUUUCCUGCUUCCCAUGGCCCCAAAAACACAAAACUUGGCCAGCGCAGAAAAUUGUUUUUCAUAUAAAUUAAUUUCCUUUUGGCGGCCAGCA------GCUAUGGGAAAAAUCCCAGGA ....(((((.((((((((((................((((((.((.((((((((((.(((......))).)))))...)))))))))))))..------))))))))))......))))) ( -31.27) >DroPer_CAF1 166963 103 - 1 UUUUUCCUGCUUCCCAUGGCCGAAAA-----AAAACC--CCCAAC--------AUGUUUU--CUAUAAAUUAAUUUCCUUUUUGUGAAAAGCAUGAAAAAGCAUGGGAAAAGUCCAAGGA ....((((((((((((((...(....-----......--..)..(--------(((((((--.((((((...........)))))).))))))))......)))))))..)))...)))) ( -21.90) >consensus CUUUUCCUGCUUCCCAUGGCCCCAAAAA__CAAAACUUGGCCAGCG_AGAAAAUUGUUUUUCAUAUAAAUUAAUUUCCUUUUGGCGGCCAGCA______GCUAUGGGAAAAAUCCCAGGA ....(((((.((((((((((...........(((((...................))))).......................((.....)).......))))))))))......))))) (-16.22 = -16.69 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:12 2006