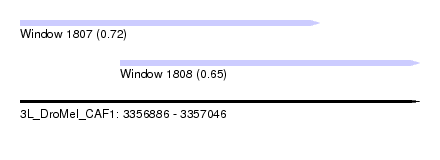
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,356,886 – 3,357,046 |
| Length | 160 |
| Max. P | 0.724458 |

| Location | 3,356,886 – 3,357,006 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -46.26 |
| Consensus MFE | -38.38 |
| Energy contribution | -38.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3356886 120 + 23771897 AGCCUCGCUGAGAUCGCUCUUGUGGCCAGCAGAAUCGUUGGCAUUGGAGGGUAUCUUGGUGCGCGACUUCUUGGCGGACGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGAUUUGACCUU ((((((((..((((((((((....((((((......))))))...)))..((((....))))))))..)))..))))..))))((((((((((((...))))))))).......)))... ( -48.31) >DroSec_CAF1 24072 120 + 1 AGCCUCGCUGAGAUCGCUCUUGUGGCUAGCAGAAUCGUUGGCAUUGGAGGAUUUUUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUU .(((.(((..((((((((((....((((((......))))))...)))(.(((....))).)))))..)))..))))))(((((..(((((((((...))))))))))))))........ ( -50.10) >DroSim_CAF1 21761 120 + 1 AGCCUCGCUGAGAUCGCUCUUGUGGCUAGCAGAAUCGUUGGCAUUGGAGGAUUUUUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUU .(((.(((..((((((((((....((((((......))))))...)))(.(((....))).)))))..)))..))))))(((((..(((((((((...))))))))))))))........ ( -50.10) >DroEre_CAF1 22634 120 + 1 AGCCUCGCUGAGAUCACUUUUGUGGCCACCAGAAUCGUUGGCGUUGGAGGGUUUCUUGGUGCGUGAUUUCUUGACGGGCGGCUGCCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUU .(((.(((..(((...(((..(..((((.(......).))))..)..)))...)))..)))(((.(.....).))))))(((.((((((((((((...))))))).)))))...).)).. ( -43.50) >DroYak_CAF1 22620 120 + 1 AGCCUCGCUGAGAUCACUUUUGUGGCCAGCAGAAUCGUUAGCAUUGGAUGGUUUCUUGGUGCGCGACUUCUUGACGGGCGGUUGUCGGUCCUCGGUAUCCGGAGAUUUGGCCUUGACCUU .((((.((((.(.((((....)))))))))((((((((..((((..(........)..)))))))).))))....))))(((((((((((..(((...)))..)).)))))...)))).. ( -39.30) >consensus AGCCUCGCUGAGAUCGCUCUUGUGGCCAGCAGAAUCGUUGGCAUUGGAGGGUUUCUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUU ((((.....((((((.((((....((((((......))))))...))))))))))...((.(((.(.....).))).))))))((((((((((((...))))))).)))))......... (-38.38 = -38.02 + -0.36)

| Location | 3,356,926 – 3,357,046 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -44.14 |
| Consensus MFE | -36.62 |
| Energy contribution | -36.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3356926 120 + 23771897 GCAUUGGAGGGUAUCUUGGUGCGCGACUUCUUGGCGGACGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGAUUUGACCUUAUCCGACGCCUCAGAUGAGGUUGUUUUCUUCGUCCCGCUA ((((..(........)..)))).........((((((..(((.((((((((((((...))))))))).((((........))))))))))...(((((((.......))))))))))))) ( -46.10) >DroSec_CAF1 24112 120 + 1 GCAUUGGAGGAUUUUUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUUAUCCGACGCCUCAGAUGAGGUUGUUUUCUUUGUCCCGCUA ((((..(((....)))..)))).........(((((((((((((..(((((((((...))))))))))))))..(((((((((.((....)).))))))))).........).))))))) ( -49.30) >DroSim_CAF1 21801 120 + 1 GCAUUGGAGGAUUUUUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUUAUCCGACGCCUCAGAUGAGGUUGUUUUCUUCGUCCCGCUA ((((..(((....)))..)))).........(((((((((((((..(((((((((...))))))))))))))..(((((((((.((....)).))))))))).........).))))))) ( -49.60) >DroEre_CAF1 22674 120 + 1 GCGUUGGAGGGUUUCUUGGUGCGUGAUUUCUUGACGGGCGGCUGCCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUUAUCAGAAGGCUCAGAUGAGCUUGUUUUCUUGGUCUCGGUG .(((..(..((((.(.......).))))..)..)))(((....)))(((((((((...)))))))))..(((..((((........((((((....))))))........))))..))). ( -40.99) >DroYak_CAF1 22660 120 + 1 GCAUUGGAUGGUUUCUUGGUGCGCGACUUCUUGACGGGCGGUUGUCGGUCCUCGGUAUCCGGAGAUUUGGCCUUGACCUUAUCGGAAGGCUCAGAUGAGGAUGUUUUUUUCGACUCGCUG ((((..(........)..))))((((..((((((.((((.(....).))))))))(((((....((((((((((..((.....))))))).)))))..)))))........)).)))).. ( -34.70) >consensus GCAUUGGAGGGUUUCUUGGUGCGCGACUUCUUGGCGGGCGGCUGUCGAUCCUCGGUAUCCGGGGAUUUGGCCUUGACCUUAUCCGACGCCUCAGAUGAGGUUGUUUUCUUCGUCCCGCUA ((((..(........)..)))).........(((((((((((((..(((((((((...))))))))))))))..(((((((((.((....)).))))))))).........).))))))) (-36.62 = -36.90 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:00:43 2006