
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,251,752 – 3,251,853 |
| Length | 101 |
| Max. P | 0.686517 |

| Location | 3,251,752 – 3,251,853 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -11.96 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
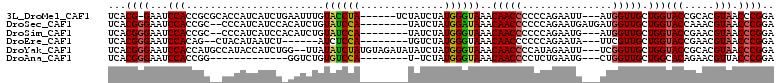
>3L_DroMel_CAF1 3251752 101 + 23771897 UCACG-GAAUCCACCGCGCACCAUCAUCUGAAUUUGUACCUA------UCUAUCUAUGGGUAAACAACCCCCCAGAAUU---AUGGUUGCUGGUACCGCACGUAACCCGGA ..(((-......((((.(((((((..((((...(((((((((------(......))))))..)))).....))))...---)))).)))))))......)))........ ( -23.00) >DroSec_CAF1 30546 101 + 1 UCACGGGAAUCCACCGC--CCCAUCAUCCACAUCUGUAUCCA--------UAUCUAUGGAUAAACAACCCCCCAGAAUGAUGAUGGUUGCUGGUACCAAACGUAACCCGGA ...((((.....(((((--.((((((((....((((((((((--------(....)))))))..........))))..))))))))..)).)))((.....))..)))).. ( -29.70) >DroSim_CAF1 33427 98 + 1 UCACGGGAAUCCACCGC--CCCAUCAUCCACAUCUGUAUCCA--------UAUCUAUGGGUAAACAACCCCCCAGAAUG---AUGGUUGCUGGUACCGAACGUAACCCGGA ...((((.....(((((--.(((((((.....((((((((((--------(....)))))))..........)))))))---))))..)).)))((.....))..)))).. ( -25.40) >DroEre_CAF1 31030 92 + 1 UCACGGGAAUCCACAG--CUACAUAAUCU------AUCUCCA--------UGUCUAUGGGUAAACAACCCCCCAGAAUA---UUCGUUGCUGGUACCGAACGUAACCCGGA ...((((......(((--(.((.......------.......--------.......((((.....))))....((...---.)))).)))).(((.....))).)))).. ( -18.50) >DroYak_CAF1 31989 106 + 1 UCACGGGAAUCCACCAUGCCAUACCAUCUGG--UUAUAUCUAUGUAGAUAUAUCUAUGGGUAAACAACCCCAUAGAAUU---UCGGUUGCUGGUACCGCACGUAACCCGGA ...((((.....((((.((...(((......--.(((((((....)))))))((((((((........))))))))...---..))).))))))((.....))..)))).. ( -31.80) >DroAna_CAF1 31812 86 + 1 UCACGGGAAUCCACCGG-------------GGUCUGUGUCCA--------U-UCUAUGGGUAAACAACCCCUCUGAAUG---CUGGUUGCUGGCACAGAACGUUACCCGGA ....((....)).((((-------------(.((((((((.(--------(-((...((((.....))))....))))(---(.....)).))))))))......))))). ( -30.30) >consensus UCACGGGAAUCCACCGC__CCCAUCAUCU_CAUCUGUAUCCA________UAUCUAUGGGUAAACAACCCCCCAGAAUG___AUGGUUGCUGGUACCGAACGUAACCCGGA ...((((...(((.......................((((((..............))))))..(((((...............))))).)))(((.....))).)))).. (-11.96 = -12.02 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:20 2006