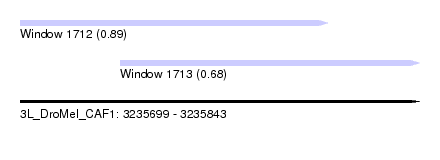
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 3,235,699 – 3,235,843 |
| Length | 144 |
| Max. P | 0.885737 |

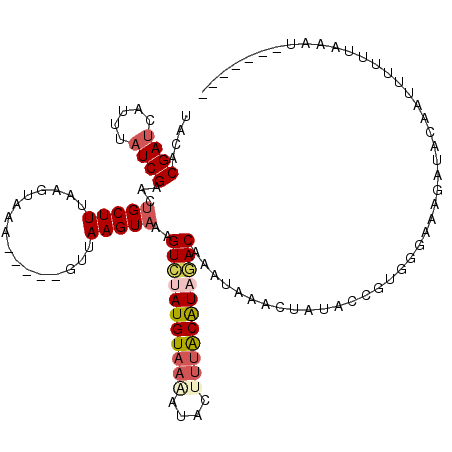
| Location | 3,235,699 – 3,235,810 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -14.51 |
| Energy contribution | -15.79 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3235699 111 + 23771897 UGUAAAAGCUUGAACAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAA-----GUUAAGUAAAGUAAAUGUAAGAUACUUUACAUAGACAAAAAA ....((((((.....))))))((((...((((((......((((.....)))).(((......))))))-----)))..((((((((.........)))))))).))))....... ( -17.90) >DroSec_CAF1 14667 116 + 1 UGUAAAAGCUUAAAAAGCUUUUCUAGGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGUAAGGUUAAGUAAGGUAUAUGUAAAAUACUUUACAUAUACAAAUAA ((((((((((.....))))))...((....))....))))......(((((..(((((((((....)))))..))))..)))))(((((((((((....)))))))))))...... ( -24.50) >DroSim_CAF1 15198 116 + 1 UGUAAAAGCUUAAAAAGCUUUUCUAGGGAACUUUCAUACACGAUCAUUUUUCGAACUGCUUUAAGUAAGGUAAGGUUAAGUAAAGUCGAUGUAAAAUACUUUACAUAGACAAAUAA ...(((((((.....)))))))..((....))....(((.(((.......)))(((((((((....)))))..))))..)))..(((.(((((((....))))))).)))...... ( -19.80) >DroEre_CAF1 14786 101 + 1 UUAAAAAGCAAGGAGAGCUUUUUUAAACUACAUC---ACACGAUCAUUUAUCGAACUGCUUUACGA--A-----CUUAAGUAAAGUCUAUGUAAAAUACA-GACGUAGAC----UA ..(((((((.......)))))))....((((.((---...((((.....))))....(((((((..--.-----.....)))))))..............-)).))))..----.. ( -18.50) >DroYak_CAF1 15059 109 + 1 UGAAAAAGCUUAAAGAGCUUUUCUAACCUACAUCAGCACACGAUCAUUUAUCGAACUGCUUUACGA--A-----UUUAAGUUAAGUCUAUGUAAAAUACCUUGCGUAGACAGCUUA ...((((((((...))))))))............((((..((((.....))))...))))......--.-----..((((((..((((((((((......)))))))))))))))) ( -24.20) >consensus UGUAAAAGCUUAAAAAGCUUUUCUAAGAAACUUUCAUACACGAUCAUUUAUCGAACUGCUUUAAGUAAA_____GUUAAGUAAAGUCUAUGUAAAAUACUUUACAUAGACAAAUAA ...(((((((.....)))))))..................((((.....))))...(((((................)))))..(((((((((((....)))))))))))...... (-14.51 = -15.79 + 1.28)

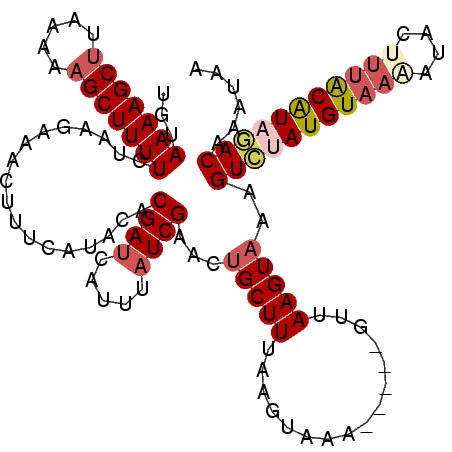
| Location | 3,235,735 – 3,235,843 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -10.35 |
| Energy contribution | -11.43 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 3235735 108 + 23771897 UACACGAUCAUUUAUCGAACUGCUUUAAGUAAA-----GUUAAGUAAAGUAAAUGUAAGAUACUUUACAUAGACAAAAAAACUACACCGUGGGAAACAUAAAAUUCUUUAAAU------- ..((((....(((((..((((...........)-----)))..)))))((..(((((((....)))))))..)).............))))(....)................------- ( -16.10) >DroSec_CAF1 14703 113 + 1 UACACGAUCAUUUAUCGAACUGCUUUAAGUAAAGUAAGGUUAAGUAAGGUAUAUGUAAAAUACUUUACAUAUACAAAUAAACUAUAGCGUGGGAAAGAUACACUUUCUUAAAU------- ..((((....(((((..(((((((((....)))))..))))..)))))(((((((((((....))))))))))).............))))((((((.....)))))).....------- ( -26.30) >DroSim_CAF1 15234 112 + 1 UACACGAUCAUUUUUCGAACUGCUUUAAGUAAGGUAAGGUUAAGUAAAGUCGAUGUAAAAUACUUUACAUAGACAAAUAAACUAUAGCGUG-GAAAGAUACAAUUUCUUAAAU------- ..((((...........(((((((((....)))))..)))).......(((.(((((((....))))))).))).............))))-..((((.......))))....------- ( -18.10) >DroEre_CAF1 14820 100 + 1 -ACACGAUCAUUUAUCGAACUGCUUUACGA--A-----CUUAAGUAAAGUCUAUGUAAAAUACA-GACGUAGAC----UAACUCUACAUUGGGAAACAUACAUUUAUUUAAAU------- -...((((.....)))).............--.-----.(((((((((((((((((........-.))))))))----)..(((......)))..........))))))))..------- ( -17.10) >DroYak_CAF1 15095 111 + 1 CACACGAUCAUUUAUCGAACUGCUUUACGA--A-----UUUAAGUUAAGUCUAUGUAAAAUACCUUGCGUAGACAGCUUAACUUUACAU-GGGAAAGAAAUUUUU-UUUAGAUAAAUAAA .........(((((((............((--(-----.(((((((..((((((((((......))))))))))))))))).)))....-((((((....)))))-)...)))))))... ( -23.20) >consensus UACACGAUCAUUUAUCGAACUGCUUUAAGUAAA_____GUUAAGUAAAGUCUAUGUAAAAUACUUUACAUAGACAAAUAAACUAUACCGUGGGAAAGAUACAAUUUUUUAAAU_______ ....((((.....))))...(((((................)))))..(((((((((((....))))))))))).............................................. (-10.35 = -11.43 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:59:12 2006