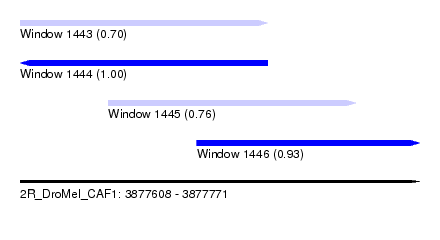
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,877,608 – 3,877,771 |
| Length | 163 |
| Max. P | 0.996906 |

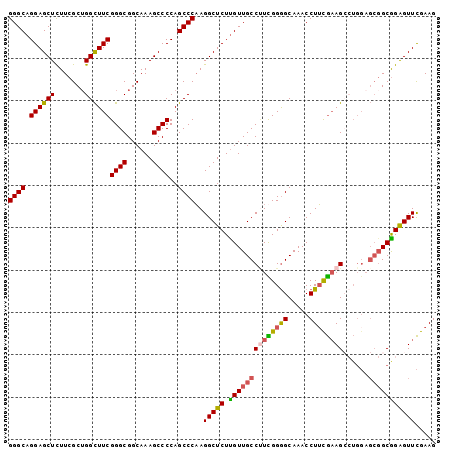
| Location | 3,877,608 – 3,877,709 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.53 |
| Mean single sequence MFE | -44.42 |
| Consensus MFE | -20.86 |
| Energy contribution | -22.70 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
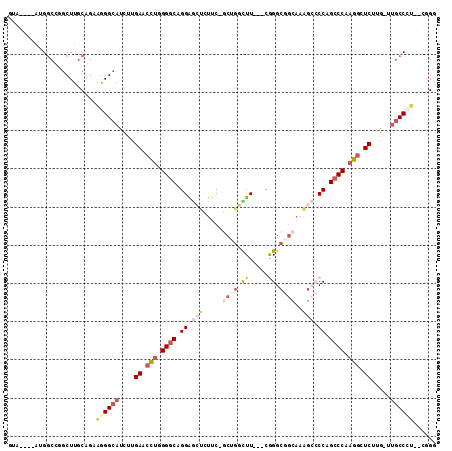
>2R_DroMel_CAF1 3877608 101 + 20766785 GUA----AUGGCCUGCUUGCAGAAGGGCAUCUUGAACCUGGGGCAGGAGCUCUUC-GCUGGCUU---CGGGCGGCAAAGCCCCAGCCCAAGGCUCUUG-UUGCCUU--UGGA .((----(.((((((....)))((((((..((((...(((((((.(((....)))-((((.(..---..).))))...)))))))..)))))))))).-..))).)--)).. ( -42.50) >DroSec_CAF1 1229 101 + 1 GUA----AUGGCCUGCUUGCAGAAGGGCAUCUUGAACCUGGGGCAGGAGCUUUUC-GCUGGUUU---CGGGCGGCAAAGCCCCAGCCCAAGGCUCUUG-UUGCCUU--UGGG ...----.....(((....)))(((((((.(..((.(((.((((.((.(((((((-(((.(...---).))))).))))).)).)))).))).))..)-.))))))--)... ( -41.80) >DroMoj_CAF1 4384 106 + 1 GGA-ACACGGCCCGGCUGGCAGCCCGGCAUUGUGACCCGUGGACAGGGAGAAUCC-UCGGUUUUCUUCAGGC---AUUGGACCAGACCUAGA-UCUGGGGCACCCAAAGUGC ...-.(((.(((.(((.....))).)))...))).((.(((....((((((((..-...))))))))....)---)).)).(((((......-))))).((((.....)))) ( -32.50) >DroAna_CAF1 1244 101 + 1 CCGGAU-AGGGCUGGCUUGCAGCGGGGCAGCUGGAGCUUAGGGCAGGAACUUUUCUGCAAUCUUUUACAGGCUG-------CCAGCCCCAAGCUCCGG-CUGCCCU--CUGC ((((..-....))))...((((.((((((((((((((((.((((((((....))))(((..((.....))..))-------)..)))).)))))))))-)))))))--)))) ( -55.80) >DroEre_CAF1 1272 101 + 1 UUG----CUGGCGGACUUGCAGAAGGGCAUCUUGAACCUGGGGCAGGAGCUCUUC-GCGGGCUU---CGGGCGGCAAAGCCCCAGCCCAAGGCUCUUG-UUGCCCU--CGGG .((----(.((....)).)))((.(((((.(..((.(((.((((.(((((((...-..))))))---)((((......))))..)))).))).))..)-.))))))--)... ( -47.00) >DroYak_CAF1 1206 101 + 1 UUG----GAGGCCGGCUUACAGAAGGGCAUCUUGAACCUGGGGCAGGAGCUCUUC-GCUGGCUU---CGGGCGGCAAAGCCCCAGCCCCAGGCUCUCG-UUGCCAU--CGGG ...----....(((((((....)))((((.(..((.((((((((.((.(((.((.-((((.(..---..).))))))))).)).)))))))).))..)-.)))).)--))). ( -46.90) >consensus GUA____AUGGCCGGCUUGCAGAAGGGCAUCUUGAACCUGGGGCAGGAGCUCUUC_GCUGGCUU___CGGGCGGCAAAGCCCCAGCCCAAGGCUCUUG_UUGCCCU__CGGG .......................((((((....((.(((.((((.((.(((.....((((.((.....)).))))..))).)).)))).))).)).....))))))...... (-20.86 = -22.70 + 1.84)


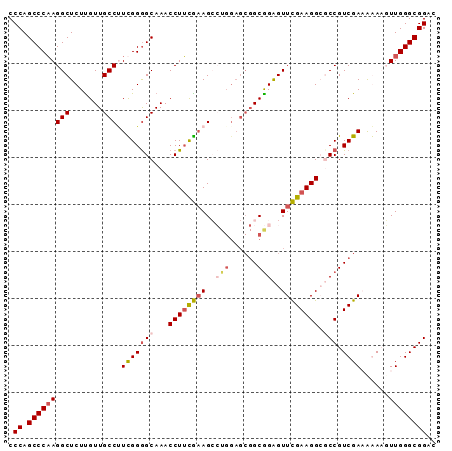
| Location | 3,877,608 – 3,877,709 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.53 |
| Mean single sequence MFE | -46.27 |
| Consensus MFE | -28.21 |
| Energy contribution | -29.38 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
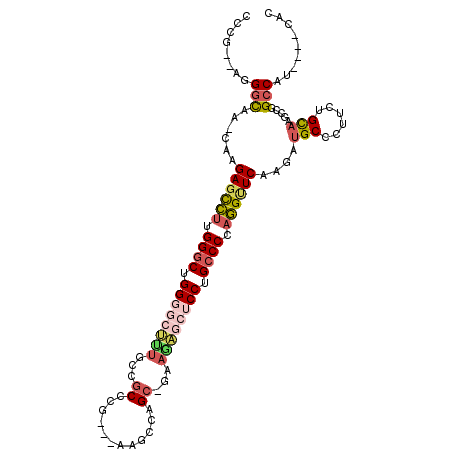
>2R_DroMel_CAF1 3877608 101 - 20766785 UCCA--AAGGCAA-CAAGAGCCUUGGGCUGGGGCUUUGCCGCCCG---AAGCCAGC-GAAGAGCUCCUGCCCCAGGUUCAAGAUGCCCUUCUGCAAGCAGGCCAU----UAC ....--..((((.-(..((((((.((((.((((((((..(((...---......))-).)))))))).)))).))))))..).))))..((((....))))....----... ( -42.80) >DroSec_CAF1 1229 101 - 1 CCCA--AAGGCAA-CAAGAGCCUUGGGCUGGGGCUUUGCCGCCCG---AAACCAGC-GAAAAGCUCCUGCCCCAGGUUCAAGAUGCCCUUCUGCAAGCAGGCCAU----UAC ....--..((((.-(..((((((.((((.((((((((..(((...---......))-).)))))))).)))).))))))..).))))..((((....))))....----... ( -42.70) >DroMoj_CAF1 4384 106 - 1 GCACUUUGGGUGCCCCAGA-UCUAGGUCUGGUCCAAU---GCCUGAAGAAAACCGA-GGAUUCUCCCUGUCCACGGGUCACAAUGCCGGGCUGCCAGCCGGGCCGUGU-UCC ...(((..((((..(((((-(....))))))..)...---)))..)))....(((.-((((.......)))).)))...(((..(((.(((.....))).)))..)))-... ( -40.00) >DroAna_CAF1 1244 101 - 1 GCAG--AGGGCAG-CCGGAGCUUGGGGCUGG-------CAGCCUGUAAAAGAUUGCAGAAAAGUUCCUGCCCUAAGCUCCAGCUGCCCCGCUGCAAGCCAGCCCU-AUCCGG ((((--.((((((-(.((((((((((((.((-------.(((((((((....))))))....))))).)))))))))))).)))))))..))))...........-...... ( -56.50) >DroEre_CAF1 1272 101 - 1 CCCG--AGGGCAA-CAAGAGCCUUGGGCUGGGGCUUUGCCGCCCG---AAGCCCGC-GAAGAGCUCCUGCCCCAGGUUCAAGAUGCCCUUCUGCAAGUCCGCCAG----CAA ...(--((((((.-(..((((((.((((.((((((((..(((...---......))-).)))))))).)))).))))))..).))))))).(((..(....)..)----)). ( -46.90) >DroYak_CAF1 1206 101 - 1 CCCG--AUGGCAA-CGAGAGCCUGGGGCUGGGGCUUUGCCGCCCG---AAGCCAGC-GAAGAGCUCCUGCCCCAGGUUCAAGAUGCCCUUCUGUAAGCCGGCCUC----CAA .(((--..((((.-(..(((((((((((.((((((((..(((...---......))-).)))))))).)))))))))))..).))))(((....))).)))....----... ( -48.70) >consensus CCCG__AGGGCAA_CAAGAGCCUUGGGCUGGGGCUUUGCCGCCCG___AAGCCAGC_GAAGAGCUCCUGCCCCAGGUUCAAGAUGCCCUUCUGCAAGCCGGCCAU____CAC ........(((......((((((.((((.((((((((...((............))...)))))))).)))).))))))....(((......))).....)))......... (-28.21 = -29.38 + 1.17)


| Location | 3,877,644 – 3,877,745 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.96 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -35.99 |
| Energy contribution | -36.17 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3877644 101 + 20766785 GGGCAGGAGCUCUUCGCUGGCUUCGGGCGGCAAAGCCCCAGCCCAAGGCUCUUGUUGCCUUUGGAGCAAACCUUCGAAGGAUGGAGCGGCAGAGUUCGAAG ......((((((((((((.((((((((((((((((((.........)))).)))))))))..)))))...((.((....)).))))))).))))))).... ( -44.80) >DroSec_CAF1 1265 92 + 1 GGGCAGGAGCUUUUCGCUGGUUUCGGGCGGCAAAGCCCCAGCCCAAGGCUCUUGUUGCCUUUGGGGCAAACCUUCAUAG---------GCGGAGUUUGAAG .((((((((((((..(((((.....(((......))))))))..))))))))))))((((.(((((.....))))).))---------))........... ( -38.40) >DroEre_CAF1 1308 101 + 1 GGGCAGGAGCUCUUCGCGGGCUUCGGGCGGCAAAGCCCCAGCCCAAGGCUCUUGUUGCCCUCGGGGCAAACCUUCGAAGCCUGGAGCGGCGGAGUUCGAAG .(((((((((.(((...(((((..((((......)))).)))))))))))))))))((((((.((((...........)))).))).)))........... ( -48.40) >DroYak_CAF1 1242 101 + 1 GGGCAGGAGCUCUUCGCUGGCUUCGGGCGGCAAAGCCCCAGCCCCAGGCUCUCGUUGCCAUCGGGGCAAACCUGUGAAGCCUGGAGCGGCGGGGUUCGAAG ((((....))))(((((((.(....).))))..((((((.(((((((((((.(((((((.....)))))...)).).)))))))...))))))))).))). ( -48.30) >consensus GGGCAGGAGCUCUUCGCUGGCUUCGGGCGGCAAAGCCCCAGCCCAAGGCUCUUGUUGCCUUCGGGGCAAACCUUCGAAGCCUGGAGCGGCGGAGUUCGAAG ((((.((((((.......))))))((((......))))..))))..(((((.((((((((((((((.....))))))))......)))))))))))..... (-35.99 = -36.17 + 0.19)

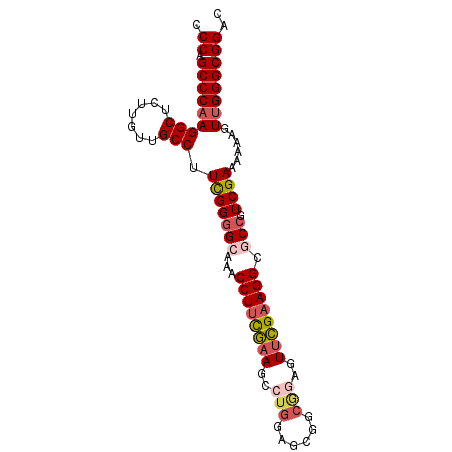
| Location | 3,877,680 – 3,877,771 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -27.36 |
| Energy contribution | -28.93 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
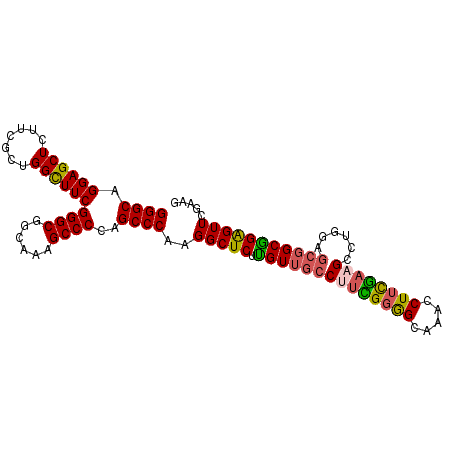
>2R_DroMel_CAF1 3877680 91 + 20766785 CCCAGCCCAAGGCUCUUGUUGCCUUUGGAGCAAACCUUCGAAGGAUGGAGCGGCAGAGUUCGAAGGCUCCGUCGAUGAAUUUUGGGCGGAC .((.((((((((((((.....(((((((((.....)))))))))..)))))(((.((((......)))).))).......))))))))).. ( -38.70) >DroSec_CAF1 1301 82 + 1 CCCAGCCCAAGGCUCUUGUUGCCUUUGGGGCAAACCUUCAUAG---------GCGGAGUUUGAAGGCUCCGUCGAUAAAUUUUGGGCGGCC ((((((((.((((.......))))...))))...........(---------((((((((....))))))))).........))))..... ( -33.60) >DroEre_CAF1 1344 91 + 1 CCCAGCCCAAGGCUCUUGUUGCCCUCGGGGCAAACCUUCGAAGCCUGGAGCGGCGGAGUUCGAAGGGGCCGUCGAAAGAAGUUGGGCGGAC .((.((((((((((....((((((...)))))).(((((((((((......)))....)))))))))))).((....))..)))))))).. ( -43.40) >DroYak_CAF1 1278 91 + 1 CCCAGCCCCAGGCUCUCGUUGCCAUCGGGGCAAACCUGUGAAGCCUGGAGCGGCGGGGUUCGAAGGGGCCGUCGAAAGAAGUUGGGCGGAC (((((((((.((..((((((((..((.((((...........)))).))))))))))..))...)))))..((....))...))))..... ( -39.20) >consensus CCCAGCCCAAGGCUCUUGUUGCCUUCGGGGCAAACCUUCGAAGCCUGGAGCGGCGGAGUUCGAAGGCGCCGUCGAAAAAAGUUGGGCGGAC .((.(((((((((.......))).(((((((...((((((((..(((......)))..)))))))).))).))))......)))))))).. (-27.36 = -28.93 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:25 2006