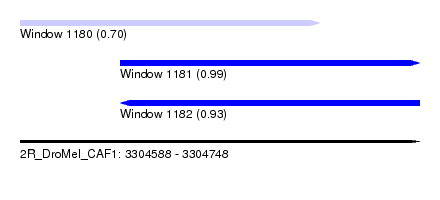
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,304,588 – 3,304,748 |
| Length | 160 |
| Max. P | 0.991082 |

| Location | 3,304,588 – 3,304,708 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -52.70 |
| Consensus MFE | -48.85 |
| Energy contribution | -48.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3304588 120 + 20766785 CUUCUUGGCGCCGCCCUUCAAGUGGGUGGACGCCUUGUCAAUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCACUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUC ......(((((.((((....(((.(.((((.(((..((...((((((.(..(((....)))..).))))))...))..))))))).))))((((((.....))..)))))))).))))). ( -52.00) >DroEre_CAF1 655 120 + 1 CUUCUUGGCACCGCCCUUCAAGUGGGUGGACGCCUUGUCGAUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUC ......(((.((((((.......))))))..)))..(((((((((((.(..(((....)))..).)))))).....)))))...(((((.(((((.(((......))).)))))))))). ( -53.20) >DroYak_CAF1 655 120 + 1 CUUCUUGGCACCGCCCUUCAAGUGGGUGGACGCCUUGUCGAUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCGGGGUCGCGUUC ......(((.(((((..((((.(((((.(.(((...(((((((((((.(..(((....)))..).)))))).....)))))....))).).))))).))))....))))).)))...... ( -52.90) >consensus CUUCUUGGCACCGCCCUUCAAGUGGGUGGACGCCUUGUCGAUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUC .....(((..((((((.......))))))..(((..((...((((((.(..(((....)))..).))))))...))..))).)))((((.(((((.(((......))).))))))))).. (-48.85 = -48.97 + 0.11)


| Location | 3,304,628 – 3,304,748 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -55.07 |
| Consensus MFE | -54.39 |
| Energy contribution | -54.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3304628 120 + 20766785 AUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCACUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUCGCUGAACACGGUGAUCUUCUGGCCGUUCACCACCAGGAAU .(((((((((((.....(((((...)))))(((((((.((((........)))).)))..)))).(((((((....(.((((((....)))))).).))))))))))))))))))..... ( -53.10) >DroEre_CAF1 695 120 + 1 AUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUCGCUGAACACGGUGAUCUUCUGGCCGUUCACCACCAGGAAU .(((((((((((.....(((((...)))))........((((...((((.(((((.(((......))).)))))))))((((((....))))))......)))))))))))))))..... ( -57.20) >DroYak_CAF1 695 120 + 1 AUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCGGGGUCGCGUUCGCUGAACACGGUGAUCUUCUGGCCGUUCACCACCAGGAAU .(((((((((((.....(((((...)))))........((((...((((.(((((.(((......))).)))))))))((((((....))))))......)))))))))))))))..... ( -54.90) >consensus AUGGUGGUGAACACUCCGGUGGACUCCACCACAUACUCGGCUCCAGCGCUGGCCCAGUUGAUGUUGGCCGGGUCGCGCUCGCUGAACACGGUGAUCUUCUGGCCGUUCACCACCAGGAAU .(((((((((((.....(((((...)))))........((((...((((.(((((.(((......))).)))))))))((((((....))))))......)))))))))))))))..... (-54.39 = -54.50 + 0.11)


| Location | 3,304,628 – 3,304,748 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -51.77 |
| Consensus MFE | -48.95 |
| Energy contribution | -49.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3304628 120 - 20766785 AUUCCUGGUGGUGAACGGCCAGAAGAUCACCGUGUUCAGCGAGCGCGACCCGGCCAACAUCAACUGGGCCAGUGCUGGAGCCGAGUAUGUGGUGGAGUCCACCGGAGUGUUCACCACCAU .....(((((((((((((((.(.....)..(((((((...)))))))....)))).((((..(((.((((.......).))).)))))))(((((...))))).....))))))))))). ( -51.00) >DroEre_CAF1 695 120 - 1 AUUCCUGGUGGUGAACGGCCAGAAGAUCACCGUGUUCAGCGAGCGCGACCCGGCCAACAUCAACUGGGCCAGCGCUGGAGCCGAGUAUGUGGUGGAGUCCACCGGAGUGUUCACCACCAU .....((((((((((((.((..........(((((((.((.(((((..(((((..........)))))...)))))...)).))))))).(((((...)))))))..)))))))))))). ( -52.30) >DroYak_CAF1 695 120 - 1 AUUCCUGGUGGUGAACGGCCAGAAGAUCACCGUGUUCAGCGAACGCGACCCCGCCAACAUCAACUGGGCCAGCGCUGGAGCCGAGUAUGUGGUGGAGUCCACCGGAGUGUUCACCACCAU .....((((((((((((.((...........((((((...))))))(((.((((((.(((..(((.((((.......).))).)))))))))))).)))....))..)))))))))))). ( -52.00) >consensus AUUCCUGGUGGUGAACGGCCAGAAGAUCACCGUGUUCAGCGAGCGCGACCCGGCCAACAUCAACUGGGCCAGCGCUGGAGCCGAGUAUGUGGUGGAGUCCACCGGAGUGUUCACCACCAU .....(((((((((((((((.(.....)..(((((((...)))))))....)))).((((..(((.((((.......).))).)))))))(((((...))))).....))))))))))). (-48.95 = -49.07 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:10 2006