
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
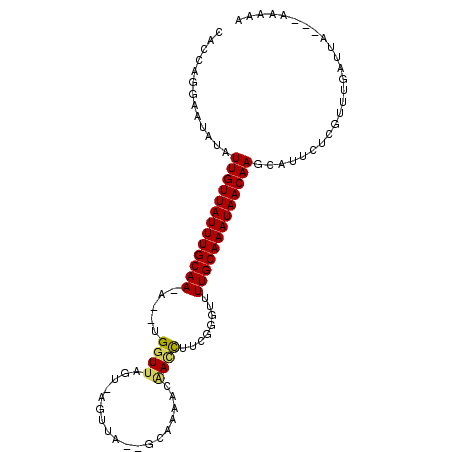
| Location | 2,979,676 – 2,979,775 |
| Length | 99 |
| Max. P | 0.977941 |

| Location | 2,979,676 – 2,979,775 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2979676 99 + 20766785 CACCAGGAAUAUAUUGUUAUUUGCAA-A--UGGUUAGU-AGUUAAGAAAAAACAACUUGCGGGUUUUGCAAAUAACAAGCAUUCUCAUUCGAUUUGAUAGAAU .....(((((...(((((((((((((-(--(.((.(((-.(((.......))).))).)).)..))))))))))))))..))))).................. ( -22.40) >DroSec_CAF1 4320 94 + 1 CACCAGGAAUAUAUUGUUAUUUGCAA-G--UGGUUAGU-AGUUG--GCAACACAACCUUCGGGUUUUGCAAAUAACAAGCAUUCUCGUUUGAUUA---AAAAA .....(((((...(((((((((((((-(--(((((((.-..)))--))..)))((((....)))))))))))))))))..)))))..........---..... ( -25.80) >DroSim_CAF1 4297 94 + 1 CACCAGGAAUAUAUUGUUAUUUGCAA-G--UGGUUAGU-AGUUG--GCAACACAACCCUCGGGUUUUGCAAAUAACAAGCAUUCUCGUUUGAUUA---AAAAA .....(((((...(((((((((((((-(--(((((((.-..)))--))..)))((((....)))))))))))))))))..)))))..........---..... ( -23.90) >DroEre_CAF1 4727 96 + 1 CACCAAGAAUAAUUUGUUAUUUGCAAAA--UGGUUAAUGAGUUA--GCAAAAACAGCUUUCAGUUUUGCAAAUAACAAGCAAACUCAUUUGAUUG---UAAAA .............(((((((((((((((--(......((((..(--((.......)))))))))))))))))))))))((((.(......).)))---).... ( -21.00) >DroYak_CAF1 4362 97 + 1 CACCGAGAAUAUUUUGUUAUUUGCAAAAUAUGGUUAAUGAGUUA--GCACA-AGAUCAUUCACUUUUGCAAAUAACAAGCCAACCCGUUUGAUUG---UAAAA ............(((((((((((((((....(((.(((((....--.....-...))))).))))))))))))))))))................---..... ( -19.40) >consensus CACCAGGAAUAUAUUGUUAUUUGCAA_A__UGGUUAGU_AGUUA__GCAAAACAACCUUCGGGUUUUGCAAAUAACAAGCAUUCUCGUUUGAUUA___AAAAA .............(((((((((((((.....((((..................))))........)))))))))))))......................... (-12.67 = -12.67 + -0.00)


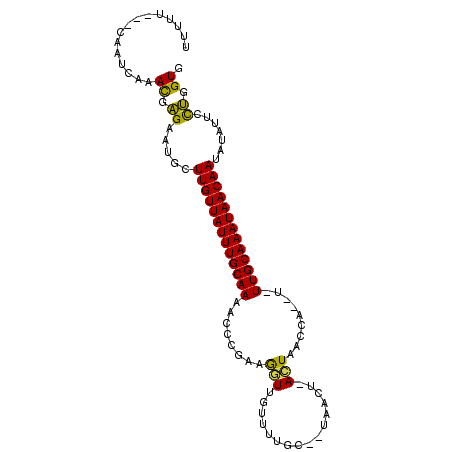
| Location | 2,979,676 – 2,979,775 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -12.00 |
| Energy contribution | -11.00 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2979676 99 - 20766785 AUUCUAUCAAAUCGAAUGAGAAUGCUUGUUAUUUGCAAAACCCGCAAGUUGUUUUUUCUUAACU-ACUAACCA--U-UUGCAAAUAACAAUAUAUUCCUGGUG ...................(((((.((((((((((((((.......(((((........)))))-........--)-)))))))))))))..)))))...... ( -18.96) >DroSec_CAF1 4320 94 - 1 UUUUU---UAAUCAAACGAGAAUGCUUGUUAUUUGCAAAACCCGAAGGUUGUGUUGC--CAACU-ACUAACCA--C-UUGCAAAUAACAAUAUAUUCCUGGUG .....---........((.(((((.(((((((((((((((((....))))((((((.--.....-..))).))--)-)))))))))))))..))))).))... ( -20.80) >DroSim_CAF1 4297 94 - 1 UUUUU---UAAUCAAACGAGAAUGCUUGUUAUUUGCAAAACCCGAGGGUUGUGUUGC--CAACU-ACUAACCA--C-UUGCAAAUAACAAUAUAUUCCUGGUG .....---........((.(((((.(((((((((((((((((....))))((((((.--.....-..))).))--)-)))))))))))))..))))).))... ( -22.70) >DroEre_CAF1 4727 96 - 1 UUUUA---CAAUCAAAUGAGUUUGCUUGUUAUUUGCAAAACUGAAAGCUGUUUUUGC--UAACUCAUUAACCA--UUUUGCAAAUAACAAAUUAUUCUUGGUG .....---..(((((..((((....(((((((((((((((.(((.(((.......))--)...))).......--)))))))))))))))...))))))))). ( -20.00) >DroYak_CAF1 4362 97 - 1 UUUUA---CAAUCAAACGGGUUGGCUUGUUAUUUGCAAAAGUGAAUGAUCU-UGUGC--UAACUCAUUAACCAUAUUUUGCAAAUAACAAAAUAUUCUCGGUG .....---........((((.((..((((((((((((((((((..((((..-.((..--..))..))))..))).)))))))))))))))..))..))))... ( -23.60) >consensus UUUUU___CAAUCAAACGAGAAUGCUUGUUAUUUGCAAAACCCGAAGGUUGUUUUGC__UAACU_ACUAACCA__U_UUGCAAAUAACAAUAUAUUCCUGGUG ...............((.((.....(((((((((((((........(((................))).........))))))))))))).......)).)). (-12.00 = -11.00 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:06 2006