| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,656,134 – 17,656,290 |
| Length | 156 |
| Max. P | 0.834691 |

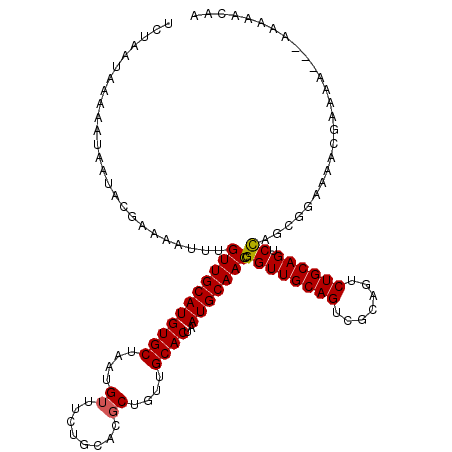
| Location | 17,656,134 – 17,656,250 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656134 116 - 20766785 GUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAG--AAAA--AAACUCAAAAUGAAACCGAAAAACAUGGAAAAAGAAAACCCC (((((((((.(((((((((....))))).)))))))).(((...(((((......)))))...)))....--....--...........))))).......................... ( -25.10) >DroSec_CAF1 9679 114 - 1 GUUUCUG----CUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAA--AAAAGCAAACUCAAAAUGAAACCGAAAAACAUGGAAAAAGAAAACCCC .((((((----((((((((....)))))).((((((((.......)))))).))))))))))........--.....................(((.......))).............. ( -25.70) >DroSim_CAF1 15626 120 - 1 GUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAACAAAAAACAAACUCAAAAUGAAACCGAAAAACAUGGAAAAAGAAAACCCC (((((((((.(((((((((....))))).)))))))).(((...(((((......)))))...))).......................))))).......................... ( -25.10) >DroEre_CAF1 9924 110 - 1 GUUUCUGCGCGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCUGUCUGCAGUCUGGCA-AAAACGAAAA------AAUAAACUCAAAAUGAAACCGAAAAACAUGGAAAAAGAAAAC--- ((((.(((.....((((((....))))))(((((((((.......)))))).))))))-.)))).....------..................(((.......)))...........--- ( -22.50) >DroYak_CAF1 21385 100 - 1 GUUUCGGAGCGCUGUUGCACUAAUGCAACCGGUUGCAG------UCUGCAGUCUAGCA-AAA-------------AAAAAUCUCAAAAUGAAACCGAAAAAAAUGGAAAAAGAAAACCCC .((((((...(((((((((....)))))).(..(((..------...)))..).))).-...-------------.....((.......))..))))))..................... ( -21.10) >consensus GUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAA___AAAAACAAACUCAAAAUGAAACCGAAAAACAUGGAAAAAGAAAACCCC .(((((.....((((((((....))))))(((((....(((...(((((......)))))...)))................((.....)))))))........))....)))))..... (-19.35 = -19.86 + 0.51)

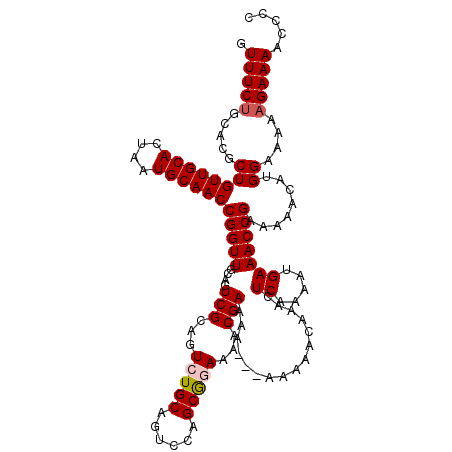

| Location | 17,656,174 – 17,656,290 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656174 116 - 20766785 UCUAAUAAAAAUAAUACGAAAAUUGGUUGCAUGUGCUAAUGUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAG--AAAA--AA (((.............(...((((((((((((((((..........))))((....))....))))))))))))...)(((...(((((......)))))...)))..))--)...--.. ( -32.30) >DroSec_CAF1 9719 114 - 1 UCUAAUAAAAAUAAUACGAAAAUUUGUUGCAUGUGCUAAUGUUUCUG----CUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAA--AAAAGCAA ......................((((((((((......))))(((((----((((((((....)))))).((((((((.......)))))).)))))))))))))))...--........ ( -29.30) >DroSim_CAF1 15666 120 - 1 UCUAAUAAAAAUAAUACGAAAAUUUGUUGCAUGUGCUAAUGUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAAACAAAAAACAA .......................(((((...(((.......(((((((.....((((((....)))))).((((((((.......)))))).)).))))))).......)))...))))) ( -30.04) >DroEre_CAF1 9961 113 - 1 UCUAAUAAAAAUAGUACGAAAAUUUGUUGCAUGUGCUAAUGUUUCUGCGCGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCUGUCUGCAGUCUGGCA-AAAACGAAAA------AAUAA ...........(((((((..((....))...))))))).(((((.(((.....((((((....))))))(((((((((.......)))))).))))))-.)))))....------..... ( -27.00) >DroYak_CAF1 21425 100 - 1 UCUAAUAAAAAUACUACGAAAAUAUGUUGCAUGUGCUAAUGUUUCGGAGCGCUGUUGCACUAAUGCAACCGGUUGCAG------UCUGCAGUCUAGCA-AAA-------------AAAAA .............((.(((((.(((...((....))..)))))))).)).(((((((((....)))))).(..(((..------...)))..).))).-...-------------..... ( -21.20) >consensus UCUAAUAAAAAUAAUACGAAAAUUUGUUGCAUGUGCUAAUGUUUCUGCACGCUGUUGCACUAAUGCAACCGGUUGCAGUCGCAGUCUGCAGUCCAGCGGAAAACGAAAA___AAAAACAA .........................(((((((((((....((........))....))))..))))))).((((((((.......)))))).)).......................... (-21.14 = -21.10 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:02 2006