
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,447,652 – 17,447,772 |
| Length | 120 |
| Max. P | 0.820753 |

| Location | 17,447,652 – 17,447,772 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
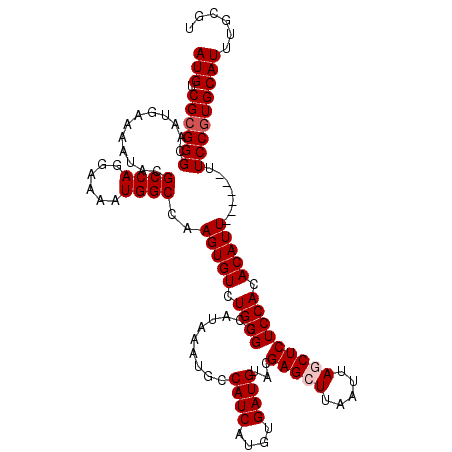
>2R_DroMel_CAF1 17447652 120 + 20766785 AUGUCGCGGGCAAUGAAAAUGCGCCAGGAAAAUGGCCAAGUGUCUGGGCCAUAAAUGUCAUCAUGUGAUGUACGAGGUUAAUUAGCUCUCCACACAUUUUCAUUUUCCGUGCAUUUGCGU (((.((((((.((((((((((.(...(((..((((((.((...)).))))))....((((((....)))).))(((.(.....).)))))).).)))))))))).)))))))))...... ( -38.50) >DroSim_CAF1 167232 120 + 1 AUGUCGCGGGCAAUGAAAAUACGCCAGGAAAAUGGCCAAGUGUCUGGGCCAUAAAUGUCAUCAUGUGAUGUACGAGGUUAAUUAGCUCUCCACACAUUUUCAUUUUCCGUGCAUUUGCGU (((.((((((.(((((((((..((((......))))...((((..((((..(((.(((((((....)))).)))...)))....))))...))))))))))))).)))))))))...... ( -36.20) >DroEre_CAF1 161237 114 + 1 AUGUCGCGGGCAAUGAAAACACGCCAGGAAAAUGGCCAAGUGUCUGGGCCAUAAAUGCCAUCAUGUGAUGUACGAGCUUAAUUAGCUCUCCACACAUU------UUCCCUGCAUUUGUGC ..(((.((((((.((....)).((((......))))....)))))))))(((((((((....(((((.((...(((((.....)))))..))))))).------......))))))))). ( -33.00) >DroYak_CAF1 147551 114 + 1 AUGUCGCGGGCAAUGAAAAUACGCCAGGAAAAUGGCCAAGUGUUUGGGCCAUAAAUGCCAUCAAAUGAUGUACGAGCUUAAUUAGCUCUCCACACAUU------UUCCGUGCAUUUGCAU (((.((((((.((((.((((((((((......))))...))))))(((..........((((....))))...(((((.....))))))))...))))------.)))))))))...... ( -32.50) >consensus AUGUCGCGGGCAAUGAAAAUACGCCAGGAAAAUGGCCAAGUGUCUGGGCCAUAAAUGCCAUCAUGUGAUGUACGAGCUUAAUUAGCUCUCCACACAUU______UUCCGUGCAUUUGCGU (((.((((((............((((......))))..(((((.((((..........((((....))))...(((((.....))))))))).))))).......)))))))))...... (-26.12 = -26.88 + 0.75)

| Location | 17,447,652 – 17,447,772 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -27.17 |
| Energy contribution | -27.55 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
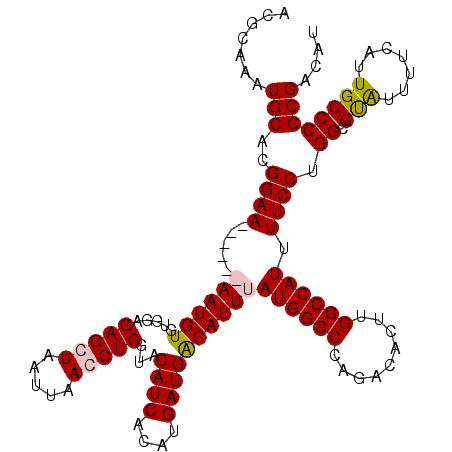
>2R_DroMel_CAF1 17447652 120 - 20766785 ACGCAAAUGCACGGAAAAUGAAAAUGUGUGGAGAGCUAAUUAACCUCGUACAUCACAUGAUGACAUUUAUGGCCCAGACACUUGGCCAUUUUCCUGGCGCAUUUUCAUUGCCCGCGACAU ......((((.(((..((((((((((((((((((.............((.((((....))))))....((((((.((...)).)))))))))))..)))))))))))))..))).).))) ( -39.80) >DroSim_CAF1 167232 120 - 1 ACGCAAAUGCACGGAAAAUGAAAAUGUGUGGAGAGCUAAUUAACCUCGUACAUCACAUGAUGACAUUUAUGGCCCAGACACUUGGCCAUUUUCCUGGCGUAUUUUCAUUGCCCGCGACAU ......((((.(((..((((((((((((((((((.............((.((((....))))))....((((((.((...)).)))))))))))..)))))))))))))..))).).))) ( -34.90) >DroEre_CAF1 161237 114 - 1 GCACAAAUGCAGGGAA------AAUGUGUGGAGAGCUAAUUAAGCUCGUACAUCACAUGAUGGCAUUUAUGGCCCAGACACUUGGCCAUUUUCCUGGCGUGUUUUCAUUGCCCGCGACAU .........(((((((------((((((((..(((((.....)))))...)).)))))...........(((((.((...)).)))))))))))))(((.((.......)).)))..... ( -35.10) >DroYak_CAF1 147551 114 - 1 AUGCAAAUGCACGGAA------AAUGUGUGGAGAGCUAAUUAAGCUCGUACAUCAUUUGAUGGCAUUUAUGGCCCAAACACUUGGCCAUUUUCCUGGCGUAUUUUCAUUGCCCGCGACAU .(((((((((.((..(------((((((((..(((((.....))))).)))).)))))..))))))))((((((.........))))))......((((.........)))).))).... ( -34.70) >consensus ACGCAAAUGCACGGAA______AAUGUGUGGAGAGCUAAUUAACCUCGUACAUCACAUGAUGACAUUUAUGGCCCAGACACUUGGCCAUUUUCCUGGCGUAUUUUCAUUGCCCGCGACAU .......(((..((((.....((((((.....(((((.....)))))...((((....))))))))))((((((.........)))))).)))).((.(((.......)))))))).... (-27.17 = -27.55 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:55 2006