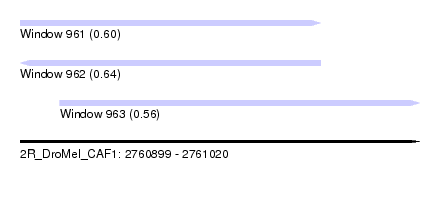
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,760,899 – 2,761,020 |
| Length | 121 |
| Max. P | 0.644160 |

| Location | 2,760,899 – 2,760,990 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -15.80 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2760899 91 + 20766785 --CAUUUCUGAUGAAAUUCAGUGGAAUUCUCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGUUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAA --.(((((....))))).(((((((.......((.(((((.((((((..........))))))..)))))..)).......)))))))..... ( -26.04) >DroVir_CAF1 50079 82 + 1 AUGCGCUCUUAUGAAAUUCG----------UUGGCGGC-GAGAGCGCAGCUAUAAAAGCGUGCGCUCGCCAGUGUCUGGCAUUCAUUGUGAAA ...(((....(((((..(((----------(.....))-))((((((((((.....))).)))))))(((((...))))).))))).)))... ( -31.10) >DroSim_CAF1 11723 91 + 1 --CAUUUCUGAUGAAAUUCAGUGGAAUUCGCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAA --.....((((......)))).....(((((((..(((((.((((((..........))))))..)))))...(....)......))))))). ( -30.40) >DroEre_CAF1 11667 91 + 1 --CAUUUCUUAUGAAAUUCAUUGGAAUUCUCAGGCGGCAGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAA --..((((....))))(((((..((((.((..((.(((((.((((((..........)))))).)).)))..))...)).))))...))))). ( -28.30) >DroYak_CAF1 11049 91 + 1 --CAUUUCUGAUGAAAUUCAGUGGAAUUCUUAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAA --.(((((....))))).(((((((...(((.((.(((((.((((((..........))))))..)))))..))..)))..)))))))..... ( -29.10) >DroPer_CAF1 10357 91 + 1 --CGUUGCUGAUGAAAUUCAGUGGGAAACGCACGUUUUGGAGAGCACUGCUAUAAAAGCGCGCGUUCGCCCGGUUGGAGCAUUCAUUGUGAAA --..(..(.((((((.((((.((((.(((((.((((((....(((...)))...)))))).)))))..)))).).)))...)))))))..).. ( -28.40) >consensus __CAUUUCUGAUGAAAUUCAGUGGAAUUCUCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAA .....((((((((((.................(((((....((((((..........))))))..)))))...(....)..))))))).))). (-15.80 = -17.17 + 1.36)

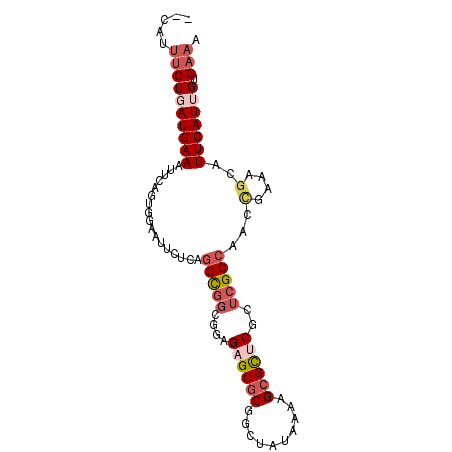
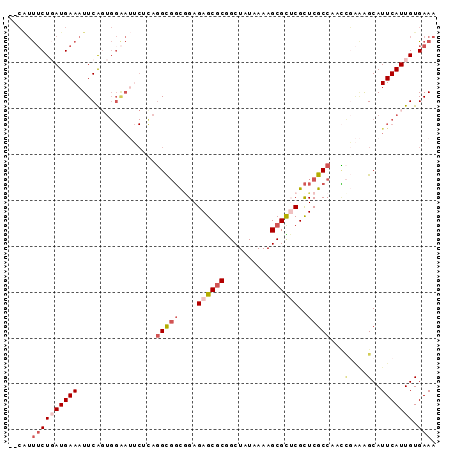
| Location | 2,760,899 – 2,760,990 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2760899 91 - 20766785 UUUCACAAUGAAUGCUUUCGGUUGGCGAGCGAACGCUUUUAUAGCCGCGCUCUCCGCCGCCUGAGAAUUCCACUGAAUUUCAUCAGAAAUG-- .........((((.(((.((((.((.(((((...(((.....)))..))))).))))))...))).))))..((((......)))).....-- ( -26.80) >DroVir_CAF1 50079 82 - 1 UUUCACAAUGAAUGCCAGACACUGGCGAGCGCACGCUUUUAUAGCUGCGCUCUC-GCCGCCAA----------CGAAUUUCAUAAGAGCGCAU .......(((((.(((((...)))))(((((((.(((.....))))))))))..-........----------.....))))).......... ( -25.90) >DroSim_CAF1 11723 91 - 1 UUUCACAAUGAAUGCUUUCGGUUGGCGAGCGAGCGCUUUUAUAGCCGCGCUCUCCGCCGCCUGCGAAUUCCACUGAAUUUCAUCAGAAAUG-- .(((.((..(((....)))(((.((((.(.((((((..........)))))).)))))))))).))).....((((......)))).....-- ( -27.60) >DroEre_CAF1 11667 91 - 1 UUUCACAAUGAAUGCUUUCGGUUGGCGAGCGAGCGCUUUUAUAGCCGCGCUCUCUGCCGCCUGAGAAUUCCAAUGAAUUUCAUAAGAAAUG-- ..(((....((((..((((((.((((.((.((((((..........)))))).)))))).))))))))))...)))(((((....))))).-- ( -27.70) >DroYak_CAF1 11049 91 - 1 UUUCACAAUGAAUGCUUUCGGUUGGCGAGCGAGCGCUUUUAUAGCCGCGCUCUCCGCCGCCUAAGAAUUCCACUGAAUUUCAUCAGAAAUG-- .........((((.(((..(((.((((.(.((((((..........)))))).)))))))).))).))))..((((......)))).....-- ( -28.30) >DroPer_CAF1 10357 91 - 1 UUUCACAAUGAAUGCUCCAACCGGGCGAACGCGCGCUUUUAUAGCAGUGCUCUCCAAAACGUGCGUUUCCCACUGAAUUUCAUCAGCAACG-- ............(((.......(((.(((((((((((.....))).((..........)))))))))))))..(((......))))))...-- ( -22.20) >consensus UUUCACAAUGAAUGCUUUCGGUUGGCGAGCGAGCGCUUUUAUAGCCGCGCUCUCCGCCGCCUGAGAAUUCCACUGAAUUUCAUCAGAAAUG__ ((((...(((((.(((.......)))(((((...(((.....)))..)))))..........................)))))..)))).... (-14.90 = -14.98 + 0.08)

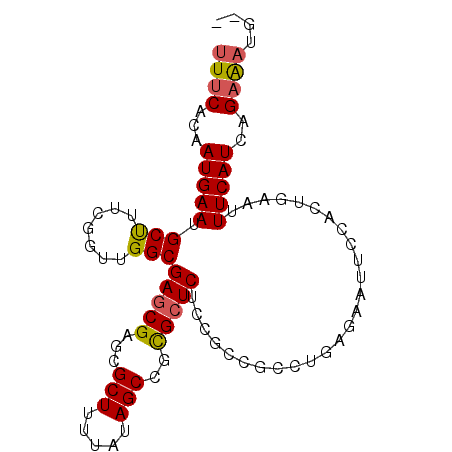
| Location | 2,760,911 – 2,761,020 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.61 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.25 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2760911 109 + 20766785 AAUUCAGUGGAAUUCUCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGUUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAAUUGCCACUCAAAAUUGAAACAG-CGCG--GACA------- .....(((((..(((.(((..(((((.((((((..........))))))..)))))...(....)......))).)))....)))))..............-....--....------- ( -27.80) >DroVir_CAF1 50093 101 + 1 AAUUCG----------UUGGCGGC-GAGAGCGCAGCUAUAAAAGCGUGCGCUCGCCAGUGUCUGGCAUUCAUUGUGAAAUUGCAAAUCAAAAUUGAAACAAGCACAAAGCAA------- ......----------...(((((-(((.((((.(((.....))))))).)))))).(((..((((((((.....)))..)))...(((....)))..))..)))...))..------- ( -33.80) >DroSim_CAF1 11735 109 + 1 AAUUCAGUGGAAUUCGCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAAUUGCCACUCAAAAUUGAAACAG-CGCG--GACA------- .....(((((..(((((((..(((((.((((((..........))))))..)))))...(....)......)))))))....)))))..............-....--....------- ( -33.80) >DroEre_CAF1 11679 109 + 1 AAUUCAUUGGAAUUCUCAGGCGGCAGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAAUUGCCACUCAAAAUUGAAACAA-CGCG--GACA------- ..(((((..((((.((..((.(((((.((((((..........)))))).)).)))..))...)).))))...))))).((((...(((....))).....-.)))--)...------- ( -27.80) >DroYak_CAF1 11061 109 + 1 AAUUCAGUGGAAUUCUUAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAAUCGCCACUCAAAAUUGAAACAA-CGCG--GACA------- ...((.(((.........((((((((.((((((..........))))))..)))))....((...(((.....)))...)))))..(((....))).....-))).--))..------- ( -31.00) >DroAna_CAF1 19421 116 + 1 AAUUCAGUGGAAUCGGUUGGUGGCGGAGAGGGCAGCUAUAAAAGCGCUCGCUCGCCACCAGGCGGCAUUCAUUGUGAAAUUGAAAAUCAAAAUUGAAACAA-CGCG--GACGACACUAG .....((((...(((.(((((((((.((.((((.(((.....))))))).)))))))))))(((...((((...(((.........)))....))))....-))).--..))))))).. ( -38.60) >consensus AAUUCAGUGGAAUUCUCAGGCGGCGGAGAGCGCGGCUAUAAAAGCGCUCGCUCGCCAACCGAAAGCAUUCAUUGUGAAAUUGCCACUCAAAAUUGAAACAA_CGCG__GACA_______ ..((((((..........((((((((.((((((..........))))))..)))))........(((.....)))......))).......))))))...................... (-21.08 = -22.25 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:29 2006