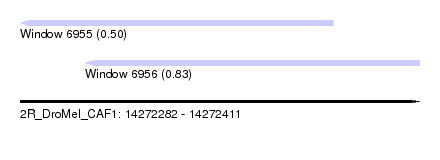
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,272,282 – 14,272,411 |
| Length | 129 |
| Max. P | 0.826635 |

| Location | 14,272,282 – 14,272,383 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.02 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.71 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
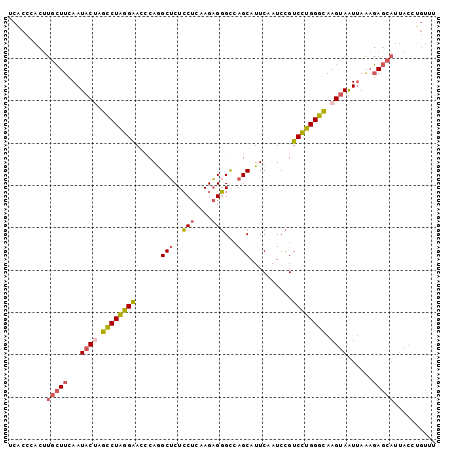
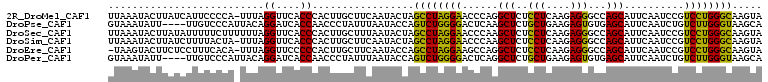
>2R_DroMel_CAF1 14272282 101 - 20766785 UCACCCACUUGCUUCAAUACUAGCCUAGGAACCCAGGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACCUGUUU .........(((((...((((.((((((((.....(((((((.....))))))).(....)......)))))))).))))......))))).......... ( -30.80) >DroSec_CAF1 1638 101 - 1 UCACCCACUUGCUUUAAUACUAGCCUAGGAACCCAAGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACCUGUUU .........((((((((((((.((((((((......((((((.....))))))..............)))))))).))).))))..))))).......... ( -28.05) >DroSim_CAF1 1638 101 - 1 UCACCCACUUGCUUCAAUACUAGCCUAGGAACCCAAGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACCUGUUU .........(((((...((((.((((((((......((((((.....))))))..............)))))))).))))......))))).......... ( -27.05) >DroEre_CAF1 1639 101 - 1 UCCCCCACUUGCUUCAAUACCAGCCUAGGAAGCCAGGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACUGGUUU ....(((..(((((...(((..((((((((.((..(((((((.....))))))).))..........))))))))..)))......)))))....)))... ( -32.40) >DroYak_CAF1 1640 101 - 1 UCACCCACUUGCUUCAAUACUAGCCUAGGAAACCAGGCGCUCCACAAGCGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACCCAUUU .........(((((...((((.((((((((......(((((((....).))))..))..........)))))))).))))......))))).......... ( -25.49) >DroPer_CAF1 3786 98 - 1 UCACCAACCCUAUUUAAUACCAGUCUGGGGACUCAGGCUCUGCUGAAGAGUGUGAGCAUUCAAUCUGUCUUGGGUAAGCAGUAAGUUA---UCUUCUGCGU ......((((..........(((..((((..((((.(((((.....))))).))))..))))..)))....))))..((((.(((...---.))))))).. ( -29.74) >consensus UCACCCACUUGCUUCAAUACUAGCCUAGGAACCCAGGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUAAUUAAAGAGCAUUACCUGUUU .........(((((...((((.((((((((......(((..(((....)))...)))..........)))))))).))))......))))).......... (-19.17 = -19.71 + 0.53)


| Location | 14,272,303 – 14,272,411 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.04 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -16.01 |
| Energy contribution | -14.46 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
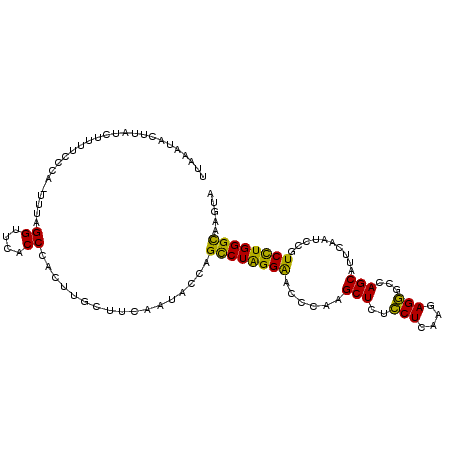
>2R_DroMel_CAF1 14272303 108 - 20766785 UUAAAUACUUAUCAUUCCCCA-UUUAGGUUCACCCACUUGCUUCAAUACUAGCCUAGGAACCCAGGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUA .....................-..(((((......)))))......((((.((((((((.....(((((((.....))))))).(....)......)))))))).)))) ( -27.50) >DroPse_CAF1 3803 105 - 1 GUAAAUAUU----UUGUCCCAUUACAGGAUCACCAACCCUAUUUAAUACCAGUCUGGGGACUCAAGCUCUGCUGAAGAGUGUGAGCAUUCAAUCUGUCUUGGGUAAGCA ........(----(((.((((..(((((........(((((.............))))).((((.(((((.....))))).)))).......)))))..)))))))).. ( -28.12) >DroSec_CAF1 1659 109 - 1 UUAAAUACUUAUAUUUUUCUUUUUUAGGUUCACCCACUUGCUUUAAUACUAGCCUAGGAACCCAAGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUA ........................(((((......)))))......((((.((((((((......((((((.....))))))..............)))))))).)))) ( -23.75) >DroSim_CAF1 1659 108 - 1 UUAAAUACUUAUCUUUUACUA-UUUAGGUUCACCCACUUGCUUCAAUACUAGCCUAGGAACCCAAGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUA .....(((((.........((-((.((((..........)))).))))...((((((((......((((((.....))))))..............))))))))))))) ( -23.95) >DroEre_CAF1 1660 107 - 1 -UAAGUACUUCUCCUUUCACA-UUUAGGUUCCCCCACUUGCUUCAAUACCAGCCUAGGAAGCCAGGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUA -(((((......(((......-...))).......)))))......(((..((((((((.((..(((((((.....))))))).))..........))))))))..))) ( -28.72) >DroPer_CAF1 3804 105 - 1 GUAAAUAUU----UUGUCCCAUUACAGGAUCACCAACCCUAUUUAAUACCAGUCUGGGGACUCAGGCUCUGCUGAAGAGUGUGAGCAUUCAAUCUGUCUUGGGUAAGCA ........(----(((.((((..(((((........(((((.............))))).((((.(((((.....))))).)))).......)))))..)))))))).. ( -30.22) >consensus UUAAAUACUUAUCUUUUCCCA_UUUAGGUUCACCCACUUGCUUCAAUACCAGCCUAGGAACCCAAGCUCUCCUCAAGAGGGCCAGCAUUCAAUCCGUCCUGGGCAAGUA ..........................((....)).................((((((((......(((..(((....)))...)))..........))))))))..... (-16.01 = -14.46 + -1.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:25 2006