
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,272,638 – 1,272,730 |
| Length | 92 |
| Max. P | 0.913419 |

| Location | 1,272,638 – 1,272,730 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1272638 92 + 20766785 AACAAUGUAUUCUUCGAUGAGCCUGCCUGAAUGAGUUUCAAAGCAGGCACAGAUACGGAAUAUACAAUAGUUUCUAUGACUGUGAUAUUUGA---------------- ..((((((((..((((.((.((((((.((((.....))))..)))))).))....))))..)))))((((((.....)))))).....))).---------------- ( -25.60) >DroGri_CAF1 4220 73 + 1 ------GUGUUCUCCGAUGAGCCUGCAUAGUUGAGUUCCAAUGCAGGAACAGAUACGGAACAUAUAAAUUCUCCACACA----------------------------- ------(((((((....((..(((((((.(........).)))))))..)).....)))))))................----------------------------- ( -18.50) >DroSec_CAF1 1233 107 + 1 AACAAUGUGUUCUUCGAUGAGCCUGCCUGAAUGAGCUUCAAAGCAGGCACAGAUACGGAAUAUACAAUAGUUUCCAUCACUGUGAUACUUGACAC-ACUAGACUCCGC .....((((((.........((((((.((((.....))))..))))))((((....((((...........))))....)))).......)))))-)........... ( -24.30) >DroSim_CAF1 1231 107 + 1 AACAAUGUGUUCUUCGAUGAGCCUGCCUGAAUGAGUUUCAAAGCAGGCACAGAUACGGAAUAUACAAUAGUUUCCAUCACUGUGAUACUUGACAC-ACUUGACUCCGC ..(((((((((.........((((((.((((.....))))..))))))((((....((((...........))))....)))).......)))))-).)))....... ( -26.70) >DroEre_CAF1 1245 108 + 1 UUCAAAAUGUUCUUCGAUGAGCCUGCCUGAUUGAGUUUCAAAGCAUGCACAGAUACGGGACAUACAAGCCAUGAAAUGAUUGUUAAACUUGACACGACUAGAUACAAC .((((.(((((((....((.((.(((.(((.......)))..))).)).)).....)))))))((((..(((...))).)))).....))))................ ( -15.70) >DroYak_CAF1 1235 108 + 1 UUUAAAAUGUUCUUCGAUGAGCCUGCCUGAUUGAGUGUCAAAGCAUGCACAGAUACGGAAUAUACAAGCCUUGAAAUCACUGUUAAAUUUGACACAACUAGAUACAAC .......(((((((((((.((.....)).)))))((((((((......((((....((..........)).((....))))))....))))))))....))).))).. ( -18.70) >consensus AACAAUGUGUUCUUCGAUGAGCCUGCCUGAAUGAGUUUCAAAGCAGGCACAGAUACGGAAUAUACAAUAGUUUCAAUCACUGUGAUACUUGACAC_ACUAGA__C__C ......(((((((....((.((((((.((((.....))))..)))))).)).....)))))))............................................. (-16.53 = -16.37 + -0.16)

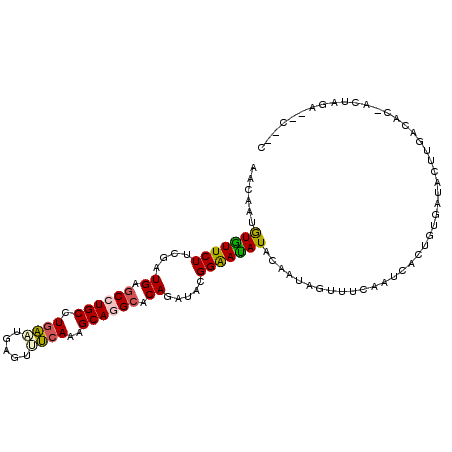
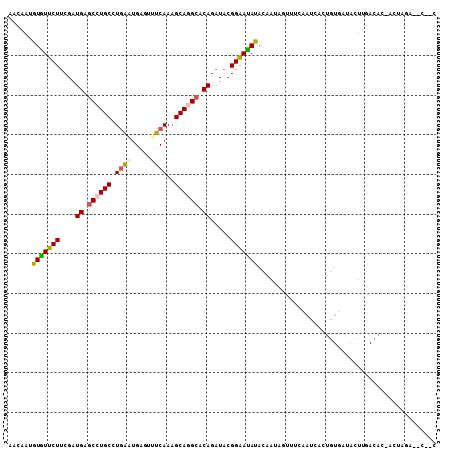
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:30 2006