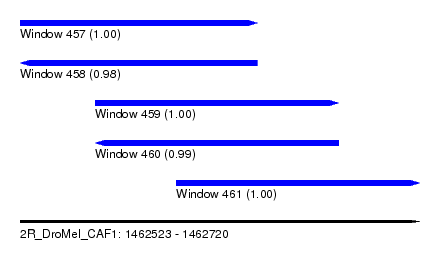
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,462,523 – 1,462,720 |
| Length | 197 |
| Max. P | 0.999950 |

| Location | 1,462,523 – 1,462,640 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -37.96 |
| Energy contribution | -38.68 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1462523 117 + 20766785 U---CAGAAGAGUAGAAAAGUUCGUUGUGCAACAAUUACAAUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCU .---......(((((...((((..(((((.......)))))..)))).....(((((((.((((..(((....))).))))))))))).((((((((((......))))))))))))))) ( -45.40) >DroSec_CAF1 83042 117 + 1 U---CAGAAGAGUAGAAAAGUUCGUUGUGCAACAAUUACAAUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGUGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCU .---......(((((...((((..(((((.......)))))..)))).....((((((((.(((..(((....))).))))))))))).((((((((((......))))))))))))))) ( -43.90) >DroSim_CAF1 82521 117 + 1 U---CAGAAGAGUAGAAAAGUUCGUUGUGCAACAAUUACAAUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCU .---......(((((...((((..(((((.......)))))..)))).....(((((((.((((..(((....))).))))))))))).((((((((((......))))))))))))))) ( -45.40) >DroEre_CAF1 94032 119 + 1 UCUCCAAAAGAGUAGAAAAGUUCGUUGUGCAGCAAUUACAAUAAGCUGGAUGGGACAACAGCUGGAGACAAAUGUCGCGGCGUUGUCCGGU-GAGUCCUGGACAAAGGACUGUGUUUGCC ...........(((((..(((((.((((.(((...((((......(.....)(((((((.((((..(((....))).))))))))))).))-))...))).)))).)))))...))))). ( -39.50) >DroYak_CAF1 89108 117 + 1 U---CAGAAGAGUAGGAAAGUUCGUUGUGCAACAAUUACAAUAAGCUAGAUGGGACAACAGCUGGAGCCGGACGUCGAGGCGUUGUCCGGCAGAGCACUGGACAAAGGUCUCUGUUUGCU .---((((((((.......(((((..((((...............((((.((......)).)))).((((((((.((...)).))))))))...)))))))))......)))).)))).. ( -34.12) >consensus U___CAGAAGAGUAGAAAAGUUCGUUGUGCAACAAUUACAAUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCU ...........(((((..((((..(((((.......)))))..)))).....(((((((.((((..(((....))).)))))))))))..(((((((((......)))))))))))))). (-37.96 = -38.68 + 0.72)


| Location | 1,462,523 – 1,462,640 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -26.26 |
| Energy contribution | -27.34 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1462523 117 - 20766785 AGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAUUGUAAUUGUUGCACAACGAACUUUUCUACUCUUCUG---A (((...(((((((((......)))))))))..(((((((((.(.(((....))).)..)).)))))))......)))........(((((....))))).................---. ( -33.50) >DroSec_CAF1 83042 117 - 1 AGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACACCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAUUGUAAUUGUUGCACAACGAACUUUUCUACUCUUCUG---A (((...(((((((((......)))))))))..((((((((.(..(((....)))....).))))))))......)))........(((((....))))).................---. ( -32.80) >DroSim_CAF1 82521 117 - 1 AGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAUUGUAAUUGUUGCACAACGAACUUUUCUACUCUUCUG---A (((...(((((((((......)))))))))..(((((((((.(.(((....))).)..)).)))))))......)))........(((((....))))).................---. ( -33.50) >DroEre_CAF1 94032 119 - 1 GGCAAACACAGUCCUUUGUCCAGGACUC-ACCGGACAACGCCGCGACAUUUGUCUCCAGCUGUUGUCCCAUCCAGCUUAUUGUAAUUGCUGCACAACGAACUUUUCUACUCUUUUGGAGA (((((..((((((((......)))))..-...(((((((((.(.(((....))).)..)).)))))))............)))..)))))..................(((.....))). ( -29.30) >DroYak_CAF1 89108 117 - 1 AGCAAACAGAGACCUUUGUCCAGUGCUCUGCCGGACAACGCCUCGACGUCCGGCUCCAGCUGUUGUCCCAUCUAGCUUAUUGUAAUUGUUGCACAACGAACUUUCCUACUCUUCUG---A ......(((((((....)))..((((...(((((((..((...))..)))))))...(((((..........))))).............))))..................))))---. ( -27.70) >consensus AGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAUUGUAAUUGUUGCACAACGAACUUUUCUACUCUUCUG___A (((...(((((((((......)))))))))..((((((((.(..(((....)))....).))))))))......)))........(((((....)))))..................... (-26.26 = -27.34 + 1.08)


| Location | 1,462,560 – 1,462,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -52.18 |
| Consensus MFE | -44.44 |
| Energy contribution | -46.20 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
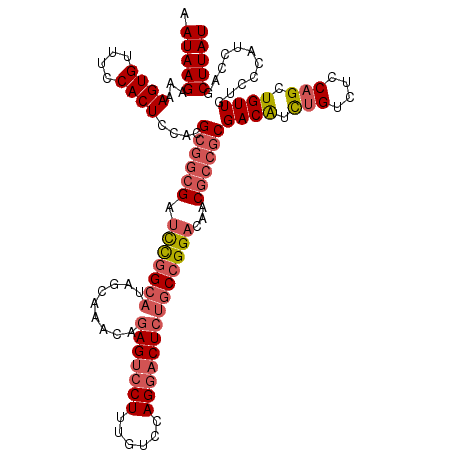
>2R_DroMel_CAF1 1462560 120 + 20766785 AUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCUUGGAGUGGAAACACUUUUCUUAUU ........(((((((..(((.(((....))).))).((((((..(((((((((((((((......))))))))(....)...)))))))))))))..((((((....))))))))))))) ( -55.70) >DroSec_CAF1 83079 120 + 1 AUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGUGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUU ........(((((((..(((.(((....))).)))(((((((..(((((((((((((((......))))))))(....)...)))))))))))))).((((((....))))))))))))) ( -55.40) >DroSim_CAF1 82558 120 + 1 AUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUU ........(((((((..(((.(((....))).)))(((((((..(((((((((((((((......))))))))(....)...)))))))))))))).((((((....))))))))))))) ( -57.30) >DroEre_CAF1 94072 119 + 1 AUAAGCUGGAUGGGACAACAGCUGGAGACAAAUGUCGCGGCGUUGUCCGGU-GAGUCCUGGACAAAGGACUGUGUUUGCCAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUU ...(((((..........)))))...(((....)))((((((..(((((((-.((((((......))))))(((.....))))))))))))))))(..(((((....)))))..)..... ( -50.40) >DroYak_CAF1 89145 120 + 1 AUAAGCUAGAUGGGACAACAGCUGGAGCCGGACGUCGAGGCGUUGUCCGGCAGAGCACUGGACAAAGGUCUCUGUUUGCUUUGCAAAUUCGUAGCAUGGAGUGGAAACACUUUUCUUAUU ....((((((..(((((((.((((....))).)((....))))))))).((((((((..((((....)))).....))))))))....)).))))..((((((....))))))....... ( -42.10) >consensus AUAAGCUGGAUGGGACAACAGCUGGAGACAGAUGUCGCGGCGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUU ........(((((((..(((.(((....))).))).((((((..(((((((((((((((......))))))))(....)...)))))))))))))..((((((....))))))))))))) (-44.44 = -46.20 + 1.76)

| Location | 1,462,560 – 1,462,680 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -30.10 |
| Energy contribution | -31.86 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
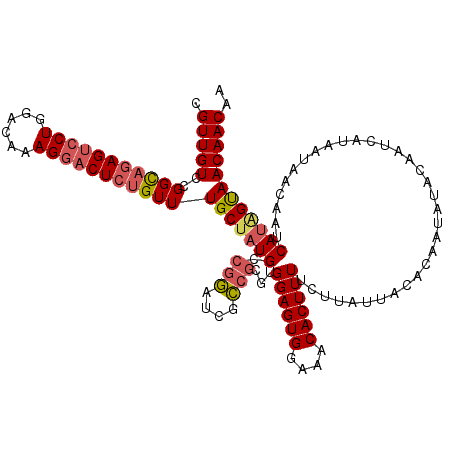
>2R_DroMel_CAF1 1462560 120 - 20766785 AAUAAGAAAAGUGUUUCCACUCCAAGCGGCGAUCCGGCAUAGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAU .........((((....))))....((((((.((((((...(....)((((((((......))))))))))))))...))))))(((....)))...(((((..........)))))... ( -42.60) >DroSec_CAF1 83079 120 - 1 AAUAAGAAAAGUGUUUCCACUCCACGCGGCGAUCCGGCAUAGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACACCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAU ....(((..((((....))))...(((((.(.((((((...(....)((((((((......))))))))))))))...).)))))...)))......(((((..........)))))... ( -38.00) >DroSim_CAF1 82558 120 - 1 AAUAAGAAAAGUGUUUCCACUCCACGCGGCGAUCCGGCAUAGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAU ....(((..((((....))))...(((((((.((((((...(....)((((((((......))))))))))))))...)))))))...)))......(((((..........)))))... ( -43.40) >DroEre_CAF1 94072 119 - 1 AAUAAGAAAAGUGUUUCCACUCCACGCGGCGAUCCGGCAUGGCAAACACAGUCCUUUGUCCAGGACUC-ACCGGACAACGCCGCGACAUUUGUCUCCAGCUGUUGUCCCAUCCAGCUUAU ....(((.((((((..........(((((((.(((((..((.....)).((((((......)))))).-.)))))...))))))))))))).)))..(((((..........)))))... ( -36.10) >DroYak_CAF1 89145 120 - 1 AAUAAGAAAAGUGUUUCCACUCCAUGCUACGAAUUUGCAAAGCAAACAGAGACCUUUGUCCAGUGCUCUGCCGGACAACGCCUCGACGUCCGGCUCCAGCUGUUGUCCCAUCUAGCUUAU .........((((....))))....((((.((....(((((((....((((..((......))..))))(((((((..((...))..)))))))....))).))))....)))))).... ( -30.40) >consensus AAUAAGAAAAGUGUUUCCACUCCACGCGGCGAUCCGGCAUAGCAAACAGAGUCCUUUGUCCAGGACUCUGCCGGACAACGCCGCGACAUCUGUCUCCAGCUGUUGUCCCAUCCAGCUUAU .(((((...((((....))))....((((((.(((((((.........(((((((......))))))))))))))...))))))((((.(((....))).))))...........))))) (-30.10 = -31.86 + 1.76)


| Location | 1,462,600 – 1,462,720 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.18 |
| SVM RNA-class probability | 0.999828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1462600 120 + 20766785 CGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCUUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUAGUAACAACAA .(((((..(((((((((((......)))))))))))(((((((.(((....)))...((((((....)))))).................................)))))))))))).. ( -38.70) >DroSec_CAF1 83119 120 + 1 UGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUGGUAACAACAA ((((((..(((((((((((......)))))))))))(((((((.(((....))).(..(((((....)))))..)...............................))))))))))))). ( -39.90) >DroSim_CAF1 82598 120 + 1 CGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUGGUAACAACAA .(((((..(((((((((((......)))))))))))(((((((.(((....))).(..(((((....)))))..)...............................)))))))))))).. ( -39.50) >DroEre_CAF1 94112 119 + 1 CGUUGUCCGGU-GAGUCCUGGACAAAGGACUGUGUUUGCCAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUAGUAACAACAA .(((((...((-(((((((......)))))(((((...((((((.((....))))))))((((....)))).........)))))....................))))....))))).. ( -36.60) >DroYak_CAF1 89185 120 + 1 CGUUGUCCGGCAGAGCACUGGACAAAGGUCUCUGUUUGCUUUGCAAAUUCGUAGCAUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUAGCAACAACAA .(((((...((((((((..((((....)))).....))))))))......((((...((((((....)))))).....))))............................)))))..... ( -29.20) >consensus CGUUGUCCGGCAGAGUCCUGGACAAAGGACUCUGUUUGCUAUGCCGGAUCGCCGCGUGGAGUGGAAACACUUUUCUUAUUACACAAUAUACAAUCAUAAUAACAAUCAUAGUAACAACAA .(((((..(((((((((((......)))))))))))(((((((.(((....)))...((((((....)))))).................................)))))))))))).. (-31.78 = -32.58 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:17 2006