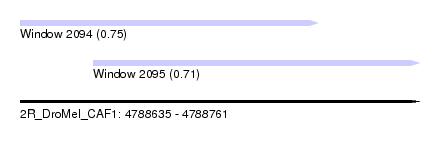
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,788,635 – 4,788,761 |
| Length | 126 |
| Max. P | 0.754651 |

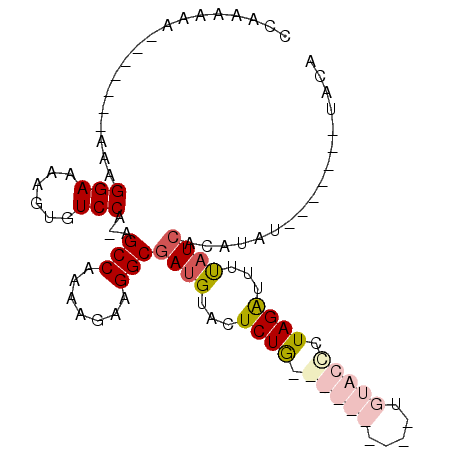
| Location | 4,788,635 – 4,788,729 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 69.77 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -9.14 |
| Energy contribution | -11.45 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4788635 94 + 20766785 CCAACAAAAAA-AAACAAGGAAAAGUGUCCAAAGCCAUAAGAAGGCGAUGUACUCUGUUGUACUCUAUGUACACUAGAUUUUAUCACAUUU-------UUCA ...........-.......(((((((((.....(((.......)))((((...((((.(((((.....))))).))))...))))))))))-------))). ( -24.40) >DroSec_CAF1 11252 94 + 1 CCAAAAAAAAACAAAAAAGGAAAAGUGUCCAAAGCCAUAAGAAGGCGAAGUACUCUG-UGUACCCUAUGUACGCUAGAUUUUAUCACAUUU-------UACA ...................(.(((((((.....(((.......)))(((((.((..(-(((((.....)))))).)))))))...))))))-------).). ( -16.30) >DroEre_CAF1 11473 84 + 1 GCAGAAAA-------AAAGGAAAAGUGUCCA-AGCCGAGAGACGGCUAUGUACUCUG----------UGUACCCUAGGCUUUAUCGCAUAUACGUAUGUAUA (((((...-------...(((......))).-(((((.....)))))......))))----------)((((.....((......))......))))..... ( -17.50) >DroYak_CAF1 11990 76 + 1 GCAGAAAA-------AAAGGAAAAGUGUCCA-AGCCAAGAAAAGGCGAUGUACUCUA----------CGUACCAUAGGUUUCAUCGCAUAU--------UUA ((......-------...(((......))).-.(((.......)))((((.((.(((----------.......)))))..))))))....--------... ( -13.70) >consensus CCAAAAAA_______AAAGGAAAAGUGUCCA_AGCCAAAAGAAGGCGAUGUACUCUG__________UGUACCCUAGAUUUUAUCACAUAU_______UACA ..................(((......)))...(((.......)))((((...((((.(((((.....))))).))))...))))................. ( -9.14 = -11.45 + 2.31)


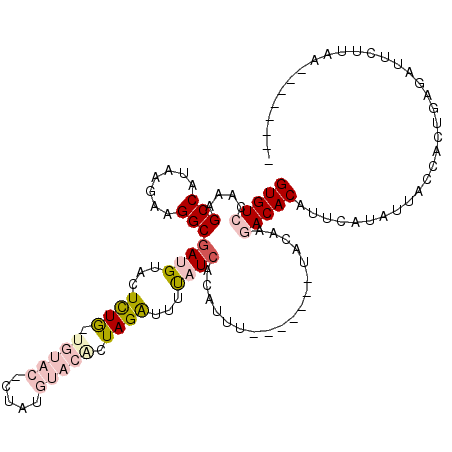
| Location | 4,788,658 – 4,788,761 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.44 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -12.38 |
| Energy contribution | -14.58 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4788658 103 + 20766785 GUGUCCAAAGCCAUAAGAAGGCGAUGUACUCUGUUGUACUCUAUGUACACUAGAUUUUAUCACAUUU-------UUCAAGACACAUUAAUAUUACCGCCGAGAUUCUUAA-------- (((((....(((.......)))((((...((((.(((((.....))))).))))...))))......-------.....)))))..........................-------- ( -23.00) >DroSec_CAF1 11276 102 + 1 GUGUCCAAAGCCAUAAGAAGGCGAAGUACUCUG-UGUACCCUAUGUACGCUAGAUUUUAUCACAUUU-------UACAACACACAUUCGUACUACCGCUGAGAUUCUUAA-------- .............((((((((((.(((((...(-(((((.....))))))..(((...)))......-------..............)))))..))))....)))))).-------- ( -18.40) >DroSim_CAF1 17982 105 + 1 GUGUCCAAAGCCAUAAGAAGGCGAAGUACUUUG-UGUACCCUAUGUACACUAGAUUUUAUCACAUUU-------UACAACACACAUUCGUAUUACGA-----AUUCCCACCACANNNN ((((.....(((.......)))(((((.((..(-(((((.....)))))).))))))).........-------....))))..(((((.....)))-----)).............. ( -19.30) >DroEre_CAF1 11490 99 + 1 GUGUCCA-AGCCGAGAGACGGCUAUGUACUCUG----------UGUACCCUAGGCUUUAUCGCAUAUACGUAUGUAUAGGACACAUACAUUUUUUCAGUAACAUUCUUAA-------- ((((((.-(((((.....))))).(((((..((----------((((......((......)).))))))...)))))))))))..........................-------- ( -25.00) >DroYak_CAF1 12007 91 + 1 GUGUCCA-AGCCAAGAAAAGGCGAUGUACUCUA----------CGUACCAUAGGUUUCAUCGCAUAU--------UUAAGACACGUAGAUAUGACCAAUUAAAUUCUUAA-------- (((((..-.(((.......)))((((.((.(((----------.......)))))..))))......--------....)))))..........................-------- ( -16.10) >consensus GUGUCCAAAGCCAUAAGAAGGCGAUGUACUCUG_UGUAC_CUAUGUACACUAGAUUUUAUCACAUUU_______UACAAGACACAUUCAUAUUACCACUGAGAUUCUUAA________ (((((....(((.......)))((((...((((.(((((.....))))).))))...))))..................))))).................................. (-12.38 = -14.58 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:54 2006