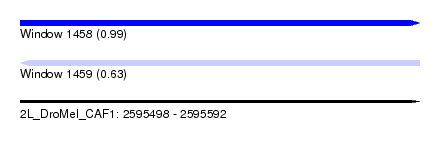
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,595,498 – 2,595,592 |
| Length | 94 |
| Max. P | 0.991217 |

| Location | 2,595,498 – 2,595,592 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.14 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
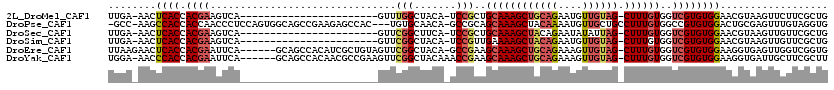
>2L_DroMel_CAF1 2595498 94 + 22407834 UUGA-AACUCACCACGAAGUCA-----------------------GUUUGGCUACA-UCCGCUGCAAAGCUGCAGAAUGUUGUAG-CUUUGUGGUCGUGUGGAACGUAAGUUCUUCGCUG ...(-((((.((......)).)-----------------------))))(((..((-..(((..(((((((((((....))))))-)))))..).))..))((((....))))...))). ( -34.10) >DroPse_CAF1 20607 114 + 1 -GCC-AAGCCACCACCAACCCUCCAGUGGCAGCCGAAGAGCCAC---UGUGCAACA-GCCGCAGCAAAGCUACAAAAUGUUGCUGCCUUUGUGGCCGUGUGGACUGCGAGUUUGUAGGUG -...-.......((((.......(((((((.........)))))---))(((((..-(((((((((..((((((((..((....)).))))))))..))....))))).))))))))))) ( -36.70) >DroSec_CAF1 16259 94 + 1 UUGA-AACUCACCACGAAGUCA-----------------------GUUCGGCUUCA-UCCGCUGCAAAGCUACAGAAUAUAUUAG-CUUUGUGGUCGUGUGGAACGUAAGUUGUUCGCUG ..((-(.....((((((((((.-----------------------....)))))).-..(((..((((((((..........)))-)))))..).)).))))(((....))).))).... ( -30.60) >DroSim_CAF1 16236 94 + 1 UUGA-AACUCACCACGAAGUCA-----------------------GUUCGGCUACA-UCCGUUGAAAAGCUACAGAAUGUUGUAG-CUUUGUGGUCGUGUGGAACGUAAGUUGUUCGCUG ....-.....(((((....(((-----------------------(..(((.....-.)))))))((((((((((....))))))-))))))))).(((.(.(((....))).).))).. ( -27.10) >DroEre_CAF1 24230 112 + 1 UUAAGAACUCACCACGAAUUCA------GCAGCCACAUCGCUGUAGUUCGGCUACA-GCCGAAGCAAAGCUGCAGAAAGUUGUAG-CUUUGUGGUCGUGUGGAAGGUGAGUUGGUCGGUG .........((((((.((((((------.(..(((((..(((((((.....)))))-))(((.((((((((((((....))))))-))))))..))))))))..).)))))).)).)))) ( -52.20) >DroYak_CAF1 16824 112 + 1 UGGA-AACCCACCACGAAUUCA------GCAGCCACAACGCCGAAGUUCGGCUACAAACCGAAGCAAAGCUGCAGAAAGUUGUAG-CUUUGUGGUCGUGUGGAAGGUGAUUGCUUCGCUU (((.-...)))...((((.(((------.(..(((((..((((.....)))).......(((.((((((((((((....))))))-))))))..))))))))..).)))....))))... ( -40.40) >consensus UUGA_AACUCACCACGAAGUCA_______________________GUUCGGCUACA_UCCGCAGCAAAGCUACAGAAUGUUGUAG_CUUUGUGGUCGUGUGGAACGUAAGUUGUUCGCUG ........((((.((((...............................(((.......)))..((((((((((((....)))))).))))))..)))))))).................. (-15.95 = -16.82 + 0.86)
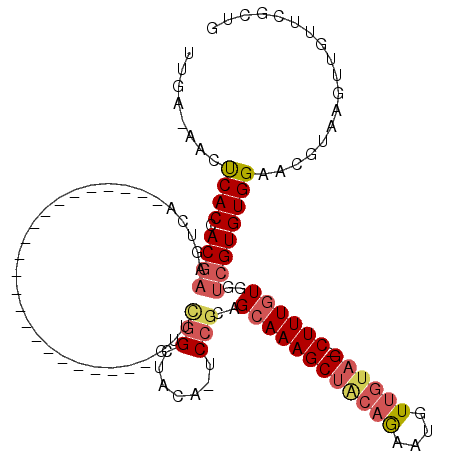
| Location | 2,595,498 – 2,595,592 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.14 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -11.23 |
| Energy contribution | -12.32 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2595498 94 - 22407834 CAGCGAAGAACUUACGUUCCACACGACCACAAAG-CUACAACAUUCUGCAGCUUUGCAGCGGA-UGUAGCCAAAC-----------------------UGACUUCGUGGUGAGUU-UCAA .......((((((.(((.....)))(((((...(-(((((.(...((((......))))..).-)))))).....-----------------------.(....)))))))))))-)... ( -24.00) >DroPse_CAF1 20607 114 - 1 CACCUACAAACUCGCAGUCCACACGGCCACAAAGGCAGCAACAUUUUGUAGCUUUGCUGCGGC-UGUUGCACA---GUGGCUCUUCGGCUGCCACUGGAGGGUUGGUGGUGGCUU-GGC- ((((.((.((((((((......((((((.(((((((.((((....)))).)))))..)).)))-)))))).((---(((((.(....)..)))))))..))))).))))))....-...- ( -46.20) >DroSec_CAF1 16259 94 - 1 CAGCGAACAACUUACGUUCCACACGACCACAAAG-CUAAUAUAUUCUGUAGCUUUGCAGCGGA-UGAAGCCGAAC-----------------------UGACUUCGUGGUGAGUU-UCAA ....(((..((((((......(((((...(((((-(((..........))))))))((((((.-.....)))..)-----------------------))...))))))))))))-)).. ( -22.40) >DroSim_CAF1 16236 94 - 1 CAGCGAACAACUUACGUUCCACACGACCACAAAG-CUACAACAUUCUGUAGCUUUUCAACGGA-UGUAGCCGAAC-----------------------UGACUUCGUGGUGAGUU-UCAA ....(((..((((.(((.....)))(((((((((-(((((......)))))))))....(((.-.....)))...-----------------------.......))))))))))-)).. ( -24.60) >DroEre_CAF1 24230 112 - 1 CACCGACCAACUCACCUUCCACACGACCACAAAG-CUACAACUUUCUGCAGCUUUGCUUCGGC-UGUAGCCGAACUACAGCGAUGUGGCUGC------UGAAUUCGUGGUGAGUUCUUAA ........((((((((..((((((((...(((((-((.((......)).)))))))..)))((-(((((.....)))))))..)))))..((------.......))))))))))..... ( -37.60) >DroYak_CAF1 16824 112 - 1 AAGCGAAGCAAUCACCUUCCACACGACCACAAAG-CUACAACUUUCUGCAGCUUUGCUUCGGUUUGUAGCCGAACUUCGGCGUUGUGGCUGC------UGAAUUCGUGGUGGGUU-UCCA ..(.(((((...((((......((((...((.((-(((((((....(((((....((....)))))))((((.....)))))))))))))..------))...)))))))).)))-))). ( -37.30) >consensus CAGCGAACAACUCACGUUCCACACGACCACAAAG_CUACAACAUUCUGCAGCUUUGCAGCGGA_UGUAGCCGAAC_______________________UGACUUCGUGGUGAGUU_UCAA ........(((((((......(((((...(((((.((.((......)).)))))))...(((.......)))...............................))))))))))))..... (-11.23 = -12.32 + 1.09)

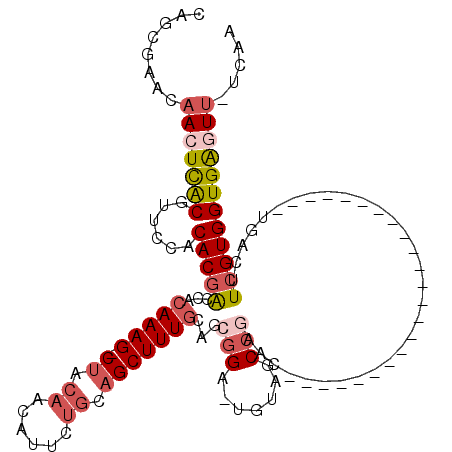
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:23 2006