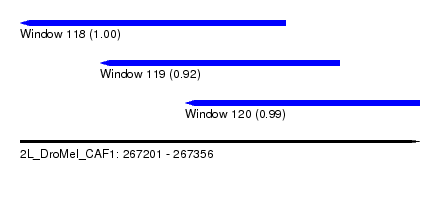
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 267,201 – 267,356 |
| Length | 155 |
| Max. P | 0.999915 |

| Location | 267,201 – 267,304 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -35.60 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 267201 103 - 22407834 CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGACUU--------AAGCGAAAAA .(((.....(((((((((.((((.....))))(((((.((.....)))))))...)))))))))...(((..(.((.........)))..)))--------..)))..... ( -31.10) >DroSec_CAF1 59001 111 - 1 CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGGCUUGAGAAUAUAAGCGAAAAA .(((......((((((((.((((.....))))(((((.((.....)))))))...))))))))(((((((..(.((.........)).).)))))))......)))..... ( -38.50) >DroSim_CAF1 72187 111 - 1 CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGACUUGAGAAUAUAAGCGAAAAA .(((......((((((((.((((.....))))(((((.((.....)))))))...))))))))(((((((..(.((.........)))..)))))))......)))..... ( -39.20) >DroEre_CAF1 65133 111 - 1 CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGACUUGAGUAUAAAAGCGAAAAA .(((.....(((((((((.((((.....))))(((((.((.....)))))))...)))))))))((((((..(.((.........)))..)))))).......)))..... ( -37.90) >DroYak_CAF1 62957 111 - 1 CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGACUUGAGUGUAAAAGCGAAAAA .(((.....(((((((((.((((.....))))(((((.((.....)))))))...)))))))))((((((..(.((.........)))..)))))).......)))..... ( -37.90) >consensus CCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGACGACAACAACAAAGUGGACUUGAGAAUAAAAGCGAAAAA .(((.....(((((((((.((((.....))))(((((.((.....)))))))...)))))))))((((((..(.((.........)))..)))))).......)))..... (-35.60 = -36.20 + 0.60)

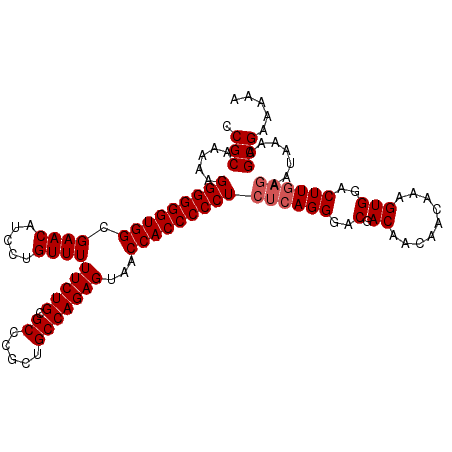
| Location | 267,232 – 267,325 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 98.71 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -29.90 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 267232 93 - 22407834 GGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...((((((....))........((((((((.((((.....))))(((((.((.....)))))))...))))))))))))..)). ( -32.50) >DroSec_CAF1 59040 93 - 1 GGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...((((((....))........((((((((.((((.....))))(((((.((.....)))))))...))))))))))))..)). ( -32.50) >DroSim_CAF1 72226 93 - 1 GGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...((((((....))........((((((((.((((.....))))(((((.((.....)))))))...))))))))))))..)). ( -32.50) >DroEre_CAF1 65172 93 - 1 GGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...((((((....))........((((((((.((((.....))))(((((.((.....)))))))...))))))))))))..)). ( -32.50) >DroYak_CAF1 62996 92 - 1 GGAAAACCGUAGCGAG-AAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...(((.(-....)))).....(((((((((.((((.....))))(((((.((.....)))))))...)))))))))....)).. ( -31.30) >consensus GGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCGCCCGCUGCCAGAGUAACCACCCCCUCUCAGGGA ......((...((((((....))........((((((((.((((.....))))(((((.((.....)))))))...))))))))))))..)). (-29.90 = -30.30 + 0.40)

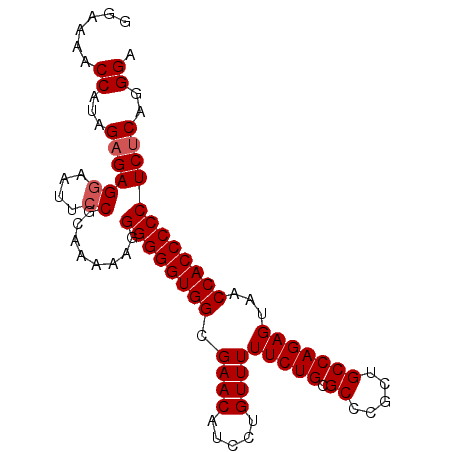
| Location | 267,265 – 267,356 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 98.24 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 267265 91 - 22407834 UGUGGGAGACAAACAAAUAGCAAAACGAGAAGGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))................((((..((.......))..))))((((((((.((((((.....)))))).))))).))). ( -22.70) >DroSec_CAF1 59073 91 - 1 UGUGGGAGACAAACAAAUAGCAAAACGAGAAGGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))................((((..((.......))..))))((((((((.((((((.....)))))).))))).))). ( -22.70) >DroSim_CAF1 72259 91 - 1 UGUGGGAGACAAACAAAUAGCAAAACGAGAAGGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))................((((..((.......))..))))((((((((.((((((.....)))))).))))).))). ( -22.70) >DroEre_CAF1 65205 91 - 1 UGUGGGAGACAAACAAAUAGCAAAACGAAAAGGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))................((((..((.......))..))))((((((((.((((((.....)))))).))))).))). ( -22.70) >DroYak_CAF1 63029 90 - 1 UGUGGGAGACAAACAAAUAGCAAAACGAGAAGGAAAACCGUAGCGAG-AAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))..........((...((((((((....(((.(-....)))).......((((((.....)))))))))))))).)) ( -21.10) >consensus UGUGGGAGACAAACAAAUAGCAAAACGAGAAGGAAAACCAUAGAGAGGAAUUCCGCAAAAAGGGGGGUGGCGAACAUCCUGUUUUUCUGCG (((.(....)..)))................((((..((.......))..))))((((((((.((((((.....)))))).))))).))). (-21.72 = -21.92 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:11 2006