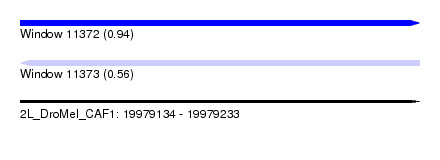
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,979,134 – 19,979,233 |
| Length | 99 |
| Max. P | 0.939226 |

| Location | 19,979,134 – 19,979,233 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
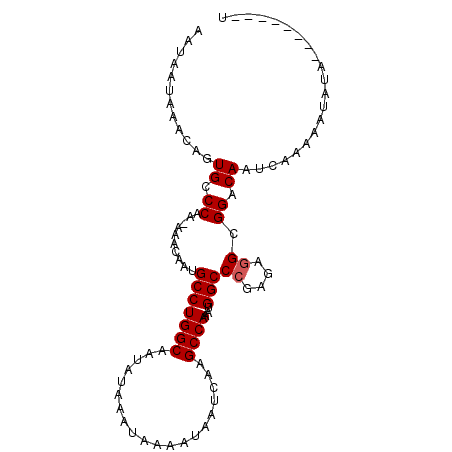
>2L_DroMel_CAF1 19979134 99 + 22407834 AUACAUACAUAUAUUUUUGAUUGUCCGCCUCUCGGGCCAUUUGGCUUGAUUAUUUUAUUUAUAUUGCCAGGCAUUGUUUUUUGGGCACUGUUUAUUAUU .....................(((((.....(((((((....)))))))...........(((.(((...))).))).....)))))............ ( -17.00) >DroSec_CAF1 162037 90 + 1 A--------UAUAUUUUUGAUUGUCCGCCUCUCGGGCCAUUUGGCUUGAUUAUUUUAUUUAUAUUGCCAGGCAUUGUUU-UUGGCCACUGUUUAUUAUU .--------.................(((..(((((((....)))))))...........(((.(((...))).)))..-..))).............. ( -14.70) >DroSim_CAF1 164574 90 + 1 A--------UAUAUUUUUGAUUGUCCGCCUCUCGGGCCAUUUGGCUUGAUUAUUUUAUUUAUAUUGCCAGGCAUUGUUU-UUGGCCACUGUUUAUUAUU .--------.................(((..(((((((....)))))))...........(((.(((...))).)))..-..))).............. ( -14.70) >DroEre_CAF1 160170 90 + 1 A--------UAUACUUUUGAUUGUCCGCCUCUCGGGCCAUUUGGCCUGAUUAUAUUAUUUAUAUUGCCAGGCAUUGUUU-UUGGGCACUCUUUAUUAUU .--------.........((.(((((((((.(((((((....))))))).((((.....)))).....)))).......-..))))).))......... ( -22.40) >DroYak_CAF1 161084 86 + 1 A--------UAUACAUUUUACUGUCCGCUUCUCGGGCCAUUUGGCUUGAUUAUUUUAUUCAUAUUGCCAGGCAUUGUUU-UUGGGCAAU----GUUAUU .--------.............(((((.....)))))....((((.(((.........)))....))))(((((((((.-...))))))----)))... ( -19.00) >consensus A________UAUAUUUUUGAUUGUCCGCCUCUCGGGCCAUUUGGCUUGAUUAUUUUAUUUAUAUUGCCAGGCAUUGUUU_UUGGGCACUGUUUAUUAUU .....................(((((.....(((((((....)))))))...........(((.(((...))).))).....)))))............ (-15.78 = -16.02 + 0.24)



| Location | 19,979,134 – 19,979,233 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -13.59 |
| Consensus MFE | -10.63 |
| Energy contribution | -10.83 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19979134 99 - 22407834 AAUAAUAAACAGUGCCCAAAAAACAAUGCCUGGCAAUAUAAAUAAAAUAAUCAAGCCAAAUGGCCCGAGAGGCGGACAAUCAAAAAUAUAUGUAUGUAU .........(((.((............)))))((((((((..............(((....)))(((.....))).............))))).))).. ( -12.80) >DroSec_CAF1 162037 90 - 1 AAUAAUAAACAGUGGCCAA-AAACAAUGCCUGGCAAUAUAAAUAAAAUAAUCAAGCCAAAUGGCCCGAGAGGCGGACAAUCAAAAAUAUA--------U ............((.((..-.......(((((((....................))))...)))((....)).)).))............--------. ( -12.35) >DroSim_CAF1 164574 90 - 1 AAUAAUAAACAGUGGCCAA-AAACAAUGCCUGGCAAUAUAAAUAAAAUAAUCAAGCCAAAUGGCCCGAGAGGCGGACAAUCAAAAAUAUA--------U ............((.((..-.......(((((((....................))))...)))((....)).)).))............--------. ( -12.35) >DroEre_CAF1 160170 90 - 1 AAUAAUAAAGAGUGCCCAA-AAACAAUGCCUGGCAAUAUAAAUAAUAUAAUCAGGCCAAAUGGCCCGAGAGGCGGACAAUCAAAAGUAUA--------U .........((.((.((..-.......((((((..((((.....))))..))))))......(((.....))))).)).)).........--------. ( -17.90) >DroYak_CAF1 161084 86 - 1 AAUAAC----AUUGCCCAA-AAACAAUGCCUGGCAAUAUGAAUAAAAUAAUCAAGCCAAAUGGCCCGAGAAGCGGACAGUAAAAUGUAUA--------U ......----((((((...-...........)))))).................(((....)))(((.....)))(((......)))...--------. ( -12.54) >consensus AAUAAUAAACAGUGCCCAA_AAACAAUGCCUGGCAAUAUAAAUAAAAUAAUCAAGCCAAAUGGCCCGAGAGGCGGACAAUCAAAAAUAUA________U ............((.((..........(((((((....................))))...)))((....)).)).))..................... (-10.63 = -10.83 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:03 2006